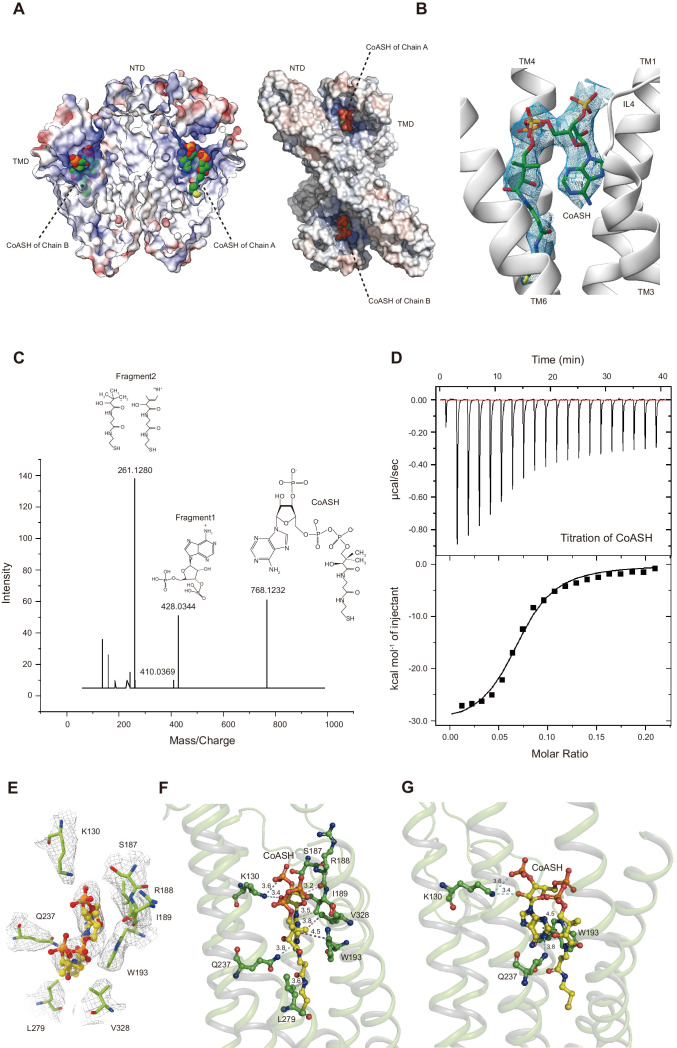
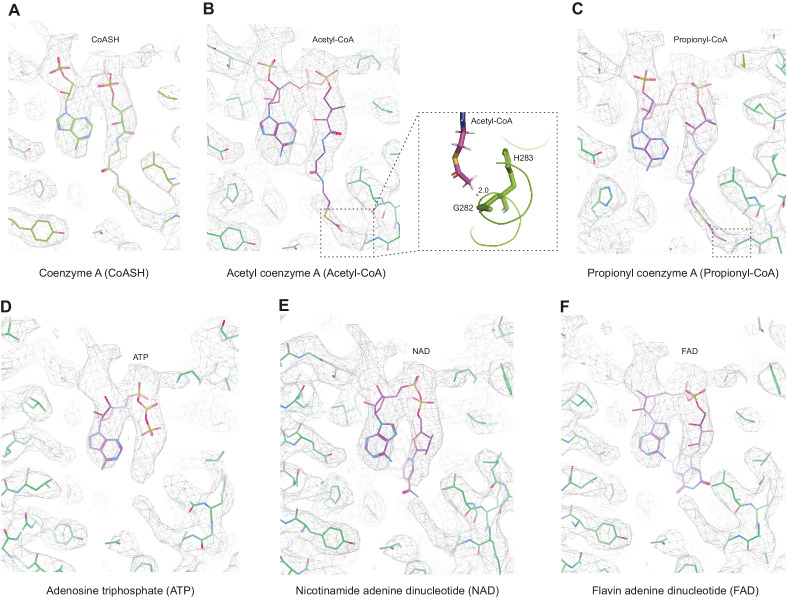
Figure 4. HsTMEM120A contains an internal CoASH-binding site within each monomer.
(A) Electrostatic potential surface presentation of HsTMEM120A dimer reveals a deep coenzyme A (CoASH)-binding cavity with an electropositive surface. Left, side view; right, top view along the membrane normal from the intracellular side. The CoASH molecules are presented as sphere models. (B) The cryo-electron microscopy (cryo-EM) density of the ligand molecule bound to HsTMEM120A fitted with a refined structural model of CoASH. (C) Mass spectrometry analysis of the small molecule extracted from the purified HsTMEM120A protein. The chemical models of CoASH molecule and its fragments are shown above the corresponding peaks with m/z values 768.1232, 428.0344, and 261.1280. (D) Isothermal titration calorimetry analysis on the kinetic interactions between CoASH and HsTMEM120A. The background heat of control was subtracted from the heat generated during binding of CoASH to HsTMEM120A. The result is fit with the single-site-binding isotherm model with ΔH = –31.09 ± 0.97 kcal/mol and Kd = 0.685 ± 0.045 μM. (E) Top view of the CoASH-binding site from the cytosolic side. The cryo-EM densities of CoASH and the surrounding amino acid residues (contoured at 9.5 rmsd) are superposed on the structural model. (F, G) Side views of the detailed interactions between CoASH and the adjacent amino acid residues of HsTMEM120A from two different angles. The blue dotted lines indicate the hydrogen bonds or the salt bridge between Lys130 NZ and CoASH O2B (3.6 Å), Lys130 NZ and CoASH O8A (3.4 Å), and Gln237 NE2 and CoASH N1A (3.8 Å). The purple dotted line shows the π-π interaction between Trp193 and CoASH at 4.2–4.5 Å distances. The pink dotted lines exhibit the non-polar interactions between CoASH and the adjacent residues (Arg188, Ile189, Leu279, and Val328) at <4 Å distance.