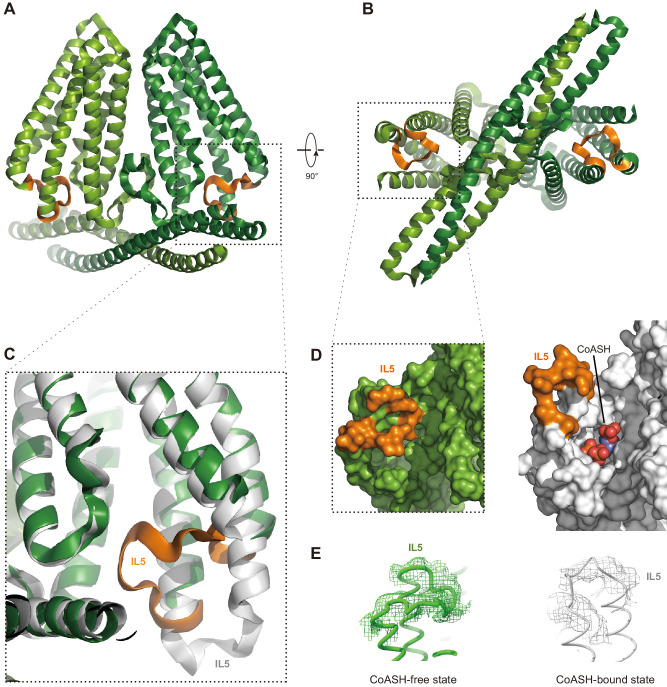
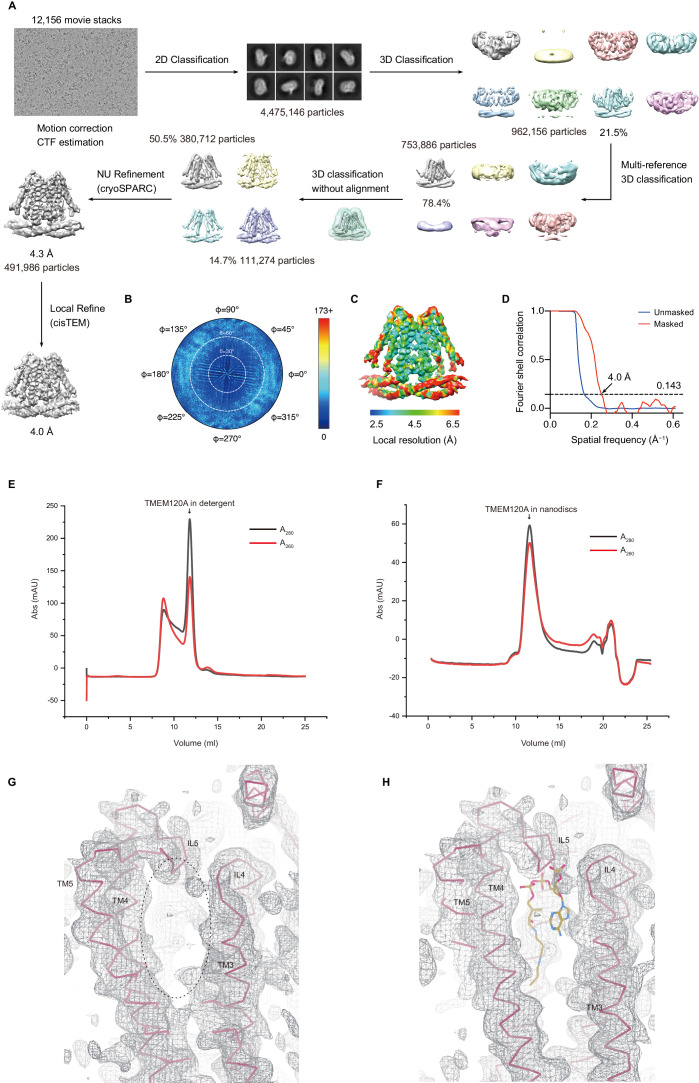
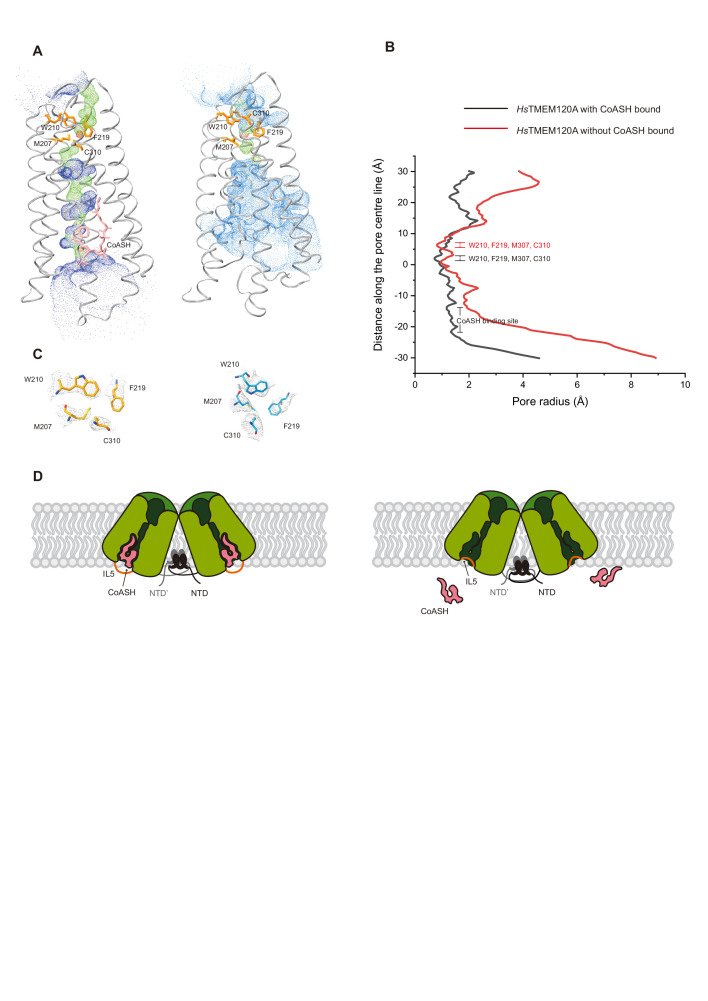
Figure 5. Structure of HsTMEM120A at the CoASH-free state in comparison with the CoASH-bound state.
(A, B) The overall structure of HsTMEM120A without coenzyme A (CoASH) bound. The side view (A) and bottom view from intracellular side (B) are shown. The two monomers are colored light and dark green, respectively, while the intracellular loop IL5 is highlighted in orange. (C) Superposition of the structures of HsTMEM120A at the CoASH-free state (green for the bulk region and orange for IL5) and CoASH-bound state (silver). The view is similar to the one in the dashed box of panel (A). (D) Surface presentation of the region around the CoASH-binding site in the CoASH-free (left) and CoASH-bound (right) HsTMEM120A structures. The IL5 loop region is highlighted in orange. (E) Cryo-electron microscopy (cryo-EM) densities of the IL5 loop in the CoASH-free (left, contoured at 4.5 rmsd) and CoASH-bound (right, contoured at 3.3 rmsd) HsTMEM120A structures.