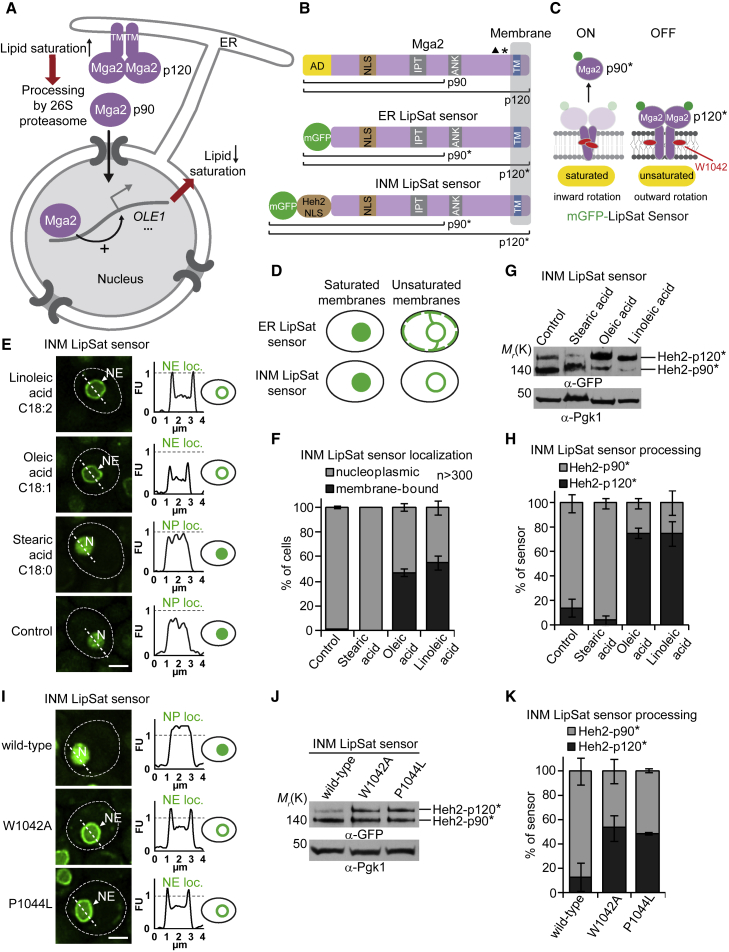
Figure 1.
The INM dynamically responds to exogenous FAs with various degree of saturation
(A) Model of the OLE1 pathway. Homodimers of Mga2 and Spt23 (not shown) are embedded in the ER as inactive precursors (p120). Transmembrane helices (TM) sense lipid saturation/unsaturation by a conformational change. When lipid saturation decreases, Mga2 becomes ubiquitinated by the E3 Rsp5, partially processed by the proteasome and is mobilized by Cdc48 (not shown). The soluble transcription factor (p90) is imported into the nucleus, where OLE1 transcription is activated.
(B) Domain organization of wild-type Mga2 and the lipid saturation (LipSat) sensors. p120 (120 kDa) designates unprocessed Mga2, p90 (90 kDa) the processed form. LipSat sensors lack the transcriptional AD and carry an N-terminal mGFP. The full-length and processed versions of the LipSat sensors are termed p120∗ and p90∗, respectively. The NLS of the INM-resident transmembrane protein Heh2 was appended to the INM LipSat sensor for nuclear import by lateral membrane diffusion. In contrast, the endogenous NLS of Mga2 promotes import of the soluble, processed p90 fragment. IPT, immunoglobulin-like/plexins/transcription factors domain required for dimerization; ANK, ankyrin repeats; TM, transmembrane domain. Triangle indicates Rsp5-binding site, asterisk depicts multiple ubiquitination sites.
(C) LipSat sensing is based on the Mga2 mechanism. A conserved tryptophane (W1042) transduces the membrane’s saturation state into an inward or outward rotational movement of the transmembrane helices. When saturated lipids increase, the sensor is activated and released from the membrane (ON). In contrast, unsaturated membranes do not trigger processing, resulting in membrane-bound LipSat sensors (OFF). Note that the E3 ligase Rsp5, the unfoldase Cdc48 and the 26S proteasome are present in both cytoplasm and nucleus allowing sensor processing in both compartments.
(D) Cartoon of predicted LipSat sensor localizations. Dashed green line beneath plasma membrane depicts peripheral ER.
(E) Live imaging of mga2Δ cells expressing the plasmid-based INM LipSat sensor supplemented with the indicated fatty acids (16 mM). Sensor fluorescence intensity was quantified across a line spanning the nucleus. For comparison the FU value 1 is marked with a horizontal dashed line. Cell contours are marked by a dashed white line. Arbitrary fluorescence units, FU; nucleus, N; nuclear envelope, NE; nucleoplasmic localization, NP loc; nuclear envelope localization, NE loc. Scale bar, 2 μm.
(F) Quantification of INM LipSat sensor localization in (E). Phenotypes were classified as membrane bound or nucleoplasmic LipSat sensor. Mean value and standard deviation are depicted. n = number of analyzed cells for each condition from 3 biological replicates.
(G) Immunoblotting analysis of INM LipSat sensor processing. Samples were taken from cell cultures used in (E). Heh2-p120∗ is membrane bound, Heh2-p90∗ is processed and soluble. Note that the GFP-tagged Heh2-p120∗/p90∗ fragments have a higher molecular weight than p120/p90. Pgk1 (3-phosphoglycerate kinase) serves as a loading control.
(H) Quantification of INM LipSat sensor processing in (G). The percentage of Heh2-p120∗ and Heh2-p90∗ relative to total amount of sensor was quantified. The mean value and standard deviation from 3 biological replicates are depicted.
(I) Live imaging of mga2Δ cells expressing the wild-type or mutant INM LipSat sensors. The conserved P1044 (aa position refers to full-length Mga2) is thought to provide conformational flexibility to the transmembrane helices during their relative rotations and facilitates the intimate interaction of two conserved W1042 residues in the dimer interface (see Figure 1C). Sensor fluorescence intensity was quantified across a line spanning the nucleus. For comparison the FU value 1 is marked with a horizontal dashed line. Arbitrary fluorescence units, FU; nucleus, N; nuclear envelope, NE; nucleoplasmic localization, NP loc; nuclear envelope localization, NE loc. Scale bar, 2 μm.
(J) Immunoblotting analysis of INM LipSat sensor processing in (I). Pgk1 serves as a loading control.
(K) Quantification of INM LipSat sensor processing in (J). The percentage of Heh2-p120∗ and Heh2-p90∗ relative to total amount of sensor was quantified. The mean value and standard deviation from 3 biological replicates are depicted.