Figure 2.
Ole1 overexpression increases UFA but not PA levels at the INM
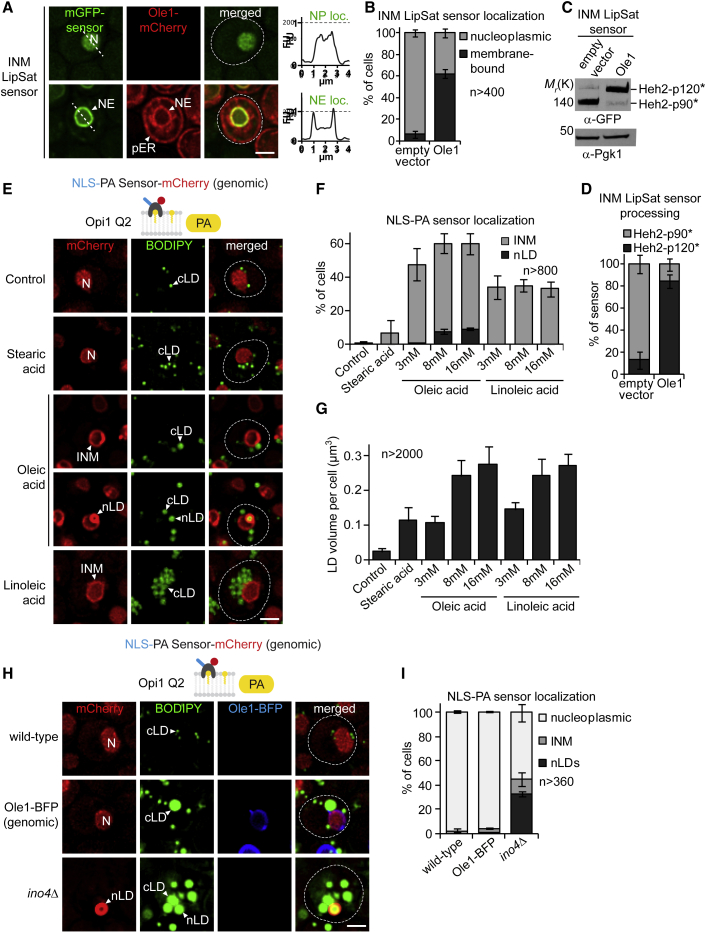
(A) Live imaging of cells expressing the INM LipSat sensor together with Ole1-mCherry (bottom panel) or an empty vector (top panel). Ole1-mCherry was expressed from the strong GPD (TDH3) promoter. Plasmids were transformed into mga2Δ cells. Sensor fluorescence intensity was quantified across a line spanning the nucleus. For comparison the FU value 1 is marked with a horizontal dashed line. Arbitrary fluorescence units, FU; nucleus, N; peripheral endoplasmic reticulum, pER; nuclear envelope, NE; nucleoplasmic localization, NP loc; nuclear envelope localization, NE loc. Scale bar, 2 μm.
(B) Quantification of INM LipSat sensor localization in (A). Phenotypes were classified as membrane bound or nucleoplasmic. Mean value and standard deviation depicted. n = number of analyzed cells for each condition from 3 biological replicates.
(C) Immunoblotting analysis of INM LipSat sensor processing in (A). Note that the GFP-tagged Heh2-p120∗/p90∗ fragments have a higher molecular weight than p120/p90. Pgk1 serves as a loading control.
(D) Quantification of INM LipSat sensor processing in (C). The percentage of Heh2-p120∗ and Heh2-p90∗ relative to total amount of sensor was quantified. The mean value and standard deviation from 3 biological replicates are depicted.
(E) Live imaging of genomically integrated NLS-PA-mCherry sensor expressed in wild-type cells, which were supplemented with the indicated fatty acids (each 16 mM dissolved in 1.5% Brij L23 solution). The NLS-PA-mCherry sensor contains the Q2 domain of the S. cerevisiae transcription factor Opi1 that specifically recognizes phosphatidic acid (PA). LDs are stained with the BODIPY dye. Nucleus, N; inner nuclear membrane, INM; nuclear lipid droplet, nLD; cytoplasmic lipid droplet, cLD. Scale bar, 2 μm.
(F) Quantification of NLS-PA-mCherry localization as observed in (E). Additional fatty acid concentrations were also quantified. n = number of analyzed cells from 3 biological replicates are depicted. Using t test, a statistically significant difference for the percentage of nLDs was verified between 16 mM oleic and 16 mM linoleic acid; and between 3 and 8 mM oleic acid. 8 and 16 mM oleic acid were not significantly different.
(G) Quantification of total LD volume per cell in (E). Additional fatty acid concentrations were also quantified. LD volumes were measured as described in STAR Methods. n = number of analyzed cells from at least 3 biological replicates. Using t test, no statistically significant difference of LD volume per cell between 16 mM oleic and 16 mM linoleic acid, or between 8 mM oleic acid and 8 mM linoleic acid was found.
(H) Live imaging of the indicated strains expressing genomically integrated NLS-PA-mCherry sensor. LDs are stained with the BODIPY dye. Genomically integrated BFP-tagged Ole1 was overexpressed from the GPD promoter. Nucleus, N; nuclear lipid droplet, nLD; cytoplasmic lipid droplet, cLD. Scale bar, 2 μm.
(I) Quantification of NLS-PA-mCherry sensor localization as observed in (H). n = number of analyzed cells from 3 biological replicates.