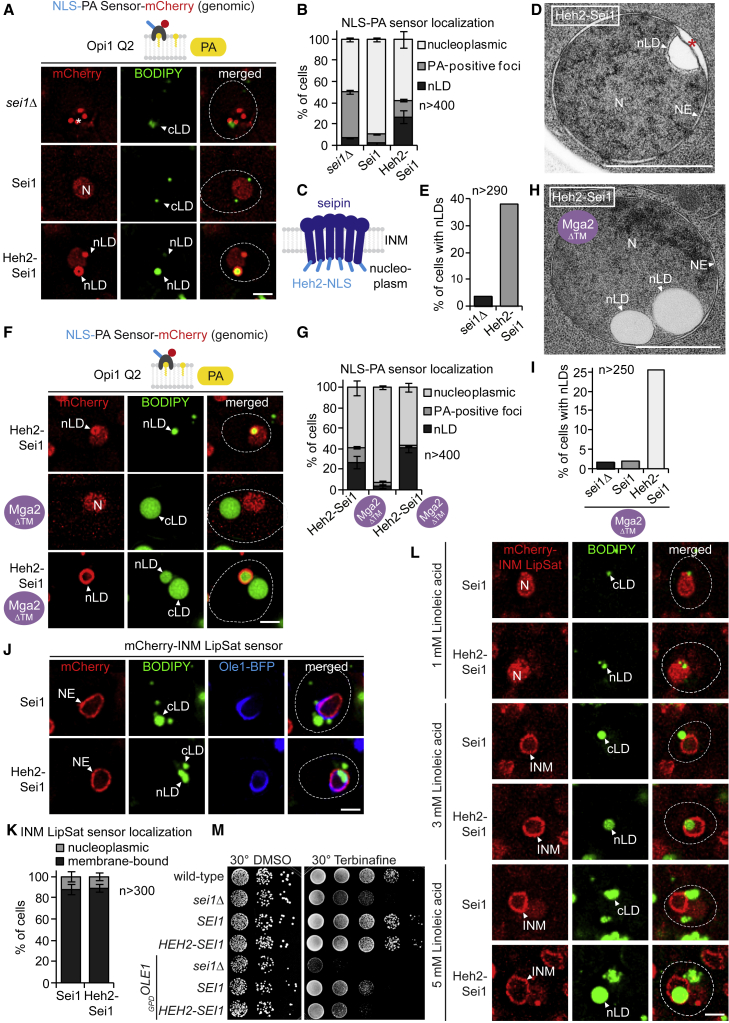
Figure 6.
Targeting seipin to the INM is sufficient to produce nLDs
(A) Live imaging of sei1Δ cells expressing the NLS-PA-mCherry sensor and the indicated SEI1 constructs or an empty vector (see also Figure S6B). SEI1 constructs were expressed from the endogenous SEI1 promoter. nLDs have a BODIPY-positive core surrounded by a PA-rich shell. Nucleus, N; nuclear lipid droplet, nLD. Asterisk marks PA-positive foci. Scale bar, 2 μm.
(B) Quantification of NLS-PA-mCherry sensor localization as observed in (A). n = number of analyzed cells obtained from 3 biological replicates.
(C) Cartoon of the engineered Sei1 structure with the Heh2-NLS attached. Putative membrane topology is based on cryo-EM models.
(D) TEM analysis of a representative example of Heh2-Sei1-expressing cells. Plasmid-based Heh2-Sei1 was expressed from the SEI1 promoter in a sei1Δ strain (see Figures S7A–S7F for a gallery). Nucleus, N; nuclear envelope, NE; nuclear lipid droplet, nLD. Asterisk marks a widened perinuclear space beneath an nLD. Scale bar, 1 μm.
(E) Quantification of nLD numbers in (D). n = number of analyzed cells.
(F) Live imaging of NLS-PA-mCherry sensor in the indicated strains as a readout for nLD production. nLDs have a BODIPY-positive core surrounded by a PA-rich shell. Nucleus, N; nuclear lipid droplet, nLD. Scale bar, 2 μm.
(G) Quantification of NLS-PA-mCherry sensor localization in (F). n = number of analyzed cells obtained from 3 biological replicates.
(H) TEM analysis of Heh2-Sei1 expression in Mga2ΔTM cells (see also Figures S7G–S7K for a gallery). Nucleus, N; nuclear envelope, NE; nuclear lipid droplet, nLD. Scale bar, 1 μm.
(I) Quantification of nLD numbers in (H). n = number of analyzed cells.
(J) Live imaging of an mCherry-tagged INM LipSat sensor in cells that overexpress genomically integrated Ole1-BFP (GPD promoter) and contain Sei1 or Heh2-Sei1 constructs. Constructs were transformed into a sei1Δ mga2Δ strain. BODIPY stains LDs. Nuclear envelope, NE; nuclear lipid droplet, nLD; cytoplasmic lipid droplet, cLD. Scale bar, 2 μm.
(K) Quantification of INM LipSat sensor localization in (J). Phenotypes were classified as membrane bound or nucleoplasmic. Mean value and standard deviation are depicted. n = number of analyzed cells for each condition from 3 biological replicates.
(L) Live imaging of an mCherry-tagged INM LipSat sensor co-expressed with Sei1 or Heh2-Sei1 constructs in cells supplemented with the indicated concentration of linoleic acid (dissolved in 1.5% Brij L23 solution). Constructs were transformed into a sei1Δ mga2Δ strain. BODIPY stains LDs. Nucleus, N; inner nuclear membrane, INM; nuclear lipid droplet, nLD; cytoplasmic lipid droplet, cLD. Scale bar, 2μm.
(M) Phenotypic analysis of the indicated strains. Genomically integrated Ole1-BFP was overexpressed from a GPD promoter. Growth was followed on SDC-LEU plates with DMSO or supplemented with 100 μg/mL Terbinafine and DMSO. Cells were spotted onto plates in 10-fold serial dilutions and incubated at 30°C.