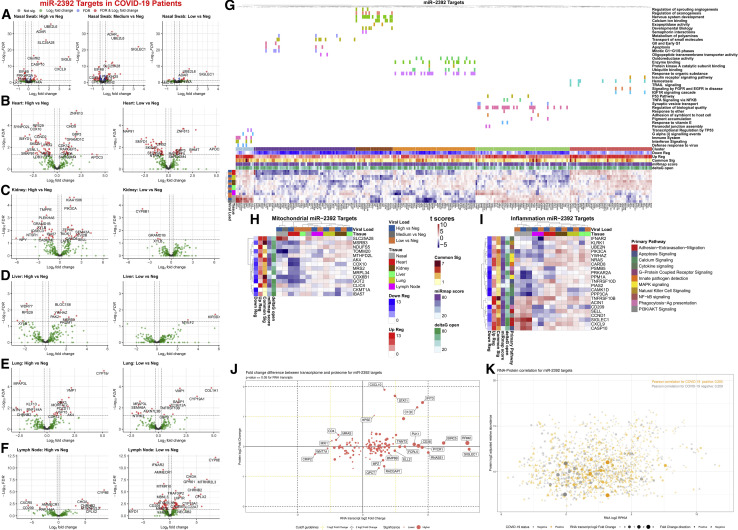
Figure 3.
Gene targets of miR-2392 in COVID-19 patients, as well as mitochondrial and inflammatory genes
(A–F) Volcano plots for differential gene expression from (A) nasopharyngeal swab and autopsy COVID-19 patient tissues from the (B) heart, (C) kidney, (D) liver, (E) lung, and (F) lymph node separated by viral load.
(G) Heatmaps displaying t score statistics when comparing viral load versus negative patient samples for nasopharyngeal swab and autopsy tissues. Main gene clusters were determined by k-means clustering and annotated using ShinyGO (Ge et al., 2020) above the heatmap.
(H and I) miR-2392 gene targets for (H) mitochondrial or (I) inflammatory genes are displayed. Genes shown have at least one comparison with a FDR < 0.05 when comparing COVID-19 patients (high, medium, or low viral loads) to non-infected patients (none). MiRmap-only heatmaps for miR-2392 mitochondrial and inflammatory gene targets are available in Figures S1 and S2, respectively.
(J) Scatterplot of log2-fold changes in RNA and protein for miR-2392 targets for genes differentially expressed at the RNA level. Student’s t test, RNA p ≤ 0.05, no limitation on the protein p value.
(K) Scatterplot of log2-transformed medians in RNA and protein. COVID-19-positive samples (orange) or COVID-19-negative samples (gray).
Student’s t test is used in fold change calculations. Shape size and opacity represent RNA fold change values (circle, positive; triangle, negative). Pearson correlations are displayed in the top corner.