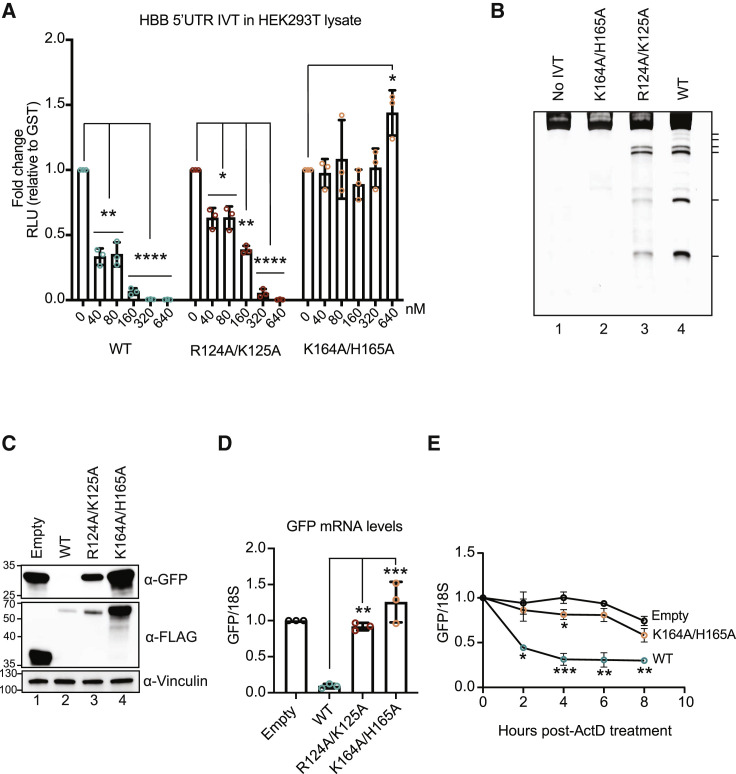
Figure 1.
CoV-2 nsp1 promotes translational suppression and mRNA decay in vitro and in cells
(A) HBB-nLuc reporter RNA was incubated with HEK293T translation extracts alone or in the presence of increasing concentrations of purified WT, R124A/K125A, or K164A/H165A nsp1. Translation of the reporter was then evaluated by luciferase assay and normalized to a glutathione S-transferase (GST) protein control. Technical triplicate measurements were taken for each biological replicate. A total of at least three biological replicates were taken for each measurement. ∗p ≤ 0.05, ∗∗p ≤ 0.01, ∗∗∗p ≤ 0.001, ∗∗∗∗p ≤ 0.0001; one-sample t test versus hypothetical value of 1. The bars represent the mean value of the replicates and error bars represent standard deviation.
(B) A primer extension assay was used to measure cleavage of the HBB-nLuc RNA in the presence of purified WT or mutant nsp1. Lane 1 (no IVT) shows nsp1 and HBB-nLuc incubation in primer extension buffer only, whereas lanes 2–4 show reactions incubated in the presence of translation extracts. Hash marks denote cleavage intermediates.
(C and D) HEK293T cells were transfected with a GFP reporter plasmid alone or together with the indicated nsp1-expressing plasmids and then harvested for protein or RNA. GFP and nsp1 protein levels were measured by ⍺-GFP and ⍺-FLAG western blots, respectively, with vinculin used as a protein loading control (C). GFP mRNA was quantified by qRT-PCR and normalized to 18S rRNA, with the level of GFP mRNA in cells lacking nsp1 then set to 1 (D). Each dot represents an independent experiment. ∗∗p ≤ 0.01, ∗∗∗p ≤ 0.001; one-way ANOVA followed by Dunnett’s multiple comparisons test versus WT nsp1. For (D), the bars represent the mean value of the replicates and error bars represent standard deviation.
(E) HEK293T cells transfected with a GFP reporter plasmid alone or together with the indicated nsp1-expressed plasmids were subsequently treated with 5-μg/mL actinomycin D (ActD) and harvested at the time points indicated after ActD treatment. GFP mRNA was quantified by qRT-PCR and normalized to 18S rRNA, and the changes in GFP mRNA abundance are relative to the time point immediately before ActD treatment. Each dot represents an independent experiment. ∗p ≤ 0.05, ∗∗p ≤ 0.01, ∗∗∗p ≤ 0.001; two-way ANOVA with Geisser-Greenhouse correction followed by Tukey’s multiple comparisons test versus “0”-h time point. See also Figures S1 and S2. The points represent the mean values of the replicates and error bars represent standard deviation.