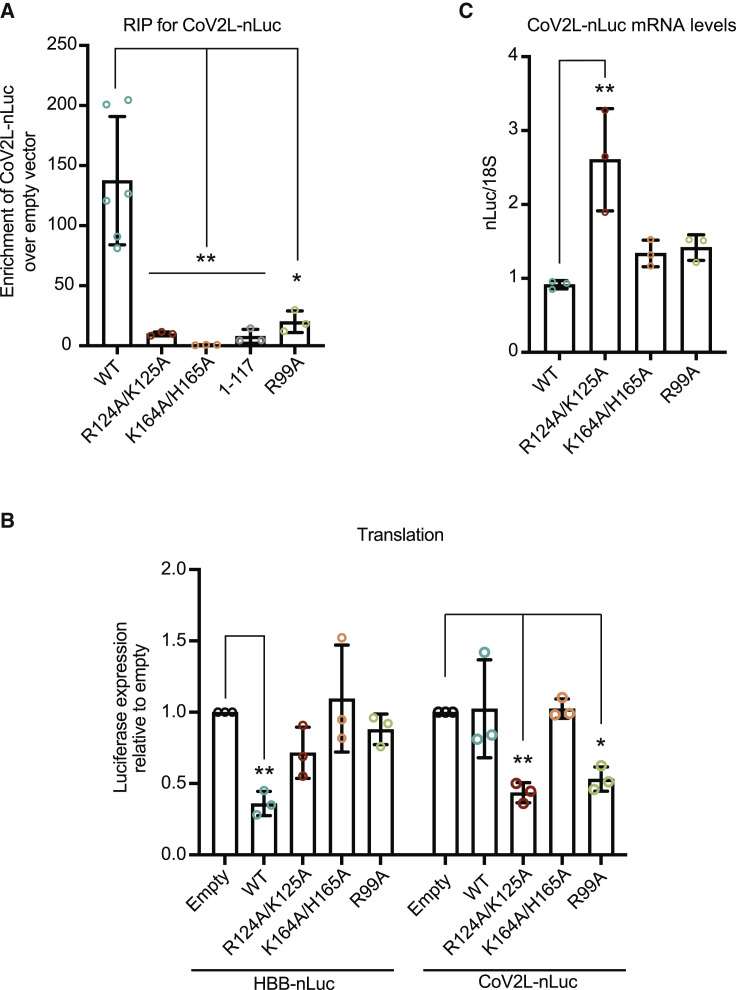
Figure 5.
Protection from translational repression conferred by the CoV-2-leader sequence is selectively eliminated by nsp1 N-terminal and central domain mutants
(A) HEK293T cells were co-transfected with a plasmid expressing CoV-2 leader-nLuc and either a control plasmid or the indicated 3xFLAG-Halo-tagged nsp1 construct. Nsp1 was immunoprecipitated using ⍺-FLAG beads, whereupon the co-immunoprecipitating RNAs were quantified by qRT-PCR. The mRNA values were then normalized to the mRNA values obtained from the empty vector control. Each dot represents an independent experiment. ∗p ≤ 0.05, ∗∗p ≤ 0.01; one-way ANOVA followed by Dunnett’s multiple comparisons test versus WT nsp1. The bars represent the mean value of the replicates and error bars represent standard deviation.
(B) HEK293T cells were transfected with either HBB-nLuc or CoV2L-nLuc together with control empty vector or the indicated nsp1 construct. Translation of HBB-nLuc or CoV2L-nLuc was measured by luciferase assay, and the fold change in luciferase activity was calculated relative to the empty vector control. Technical triplicate measurements were taken for each biological replicate. A total of at least three biological replicates were taken for each measurement. ∗p ≤ 0.05, ∗∗p ≤ 0.01; one-sample t test versus hypothetical value of 1. The bars represent the mean value of the replicates and error bars represent standard deviation.
(C) CoV2L-nLuc mRNA was quantified from the above experiment by qRT-PCR and normalized to 18S rRNA, with the level of CoV2L-nLuc mRNA in cells lacking nsp1 then set to 1. Each dot represents an independent experiment. ∗∗p ≤ 0.01; one-way ANOVA followed by Dunnett’s multiple comparisons test versus WT nsp1. See also Figure S6. The bars represent the mean value of the replicates and error bars represent standard deviation.