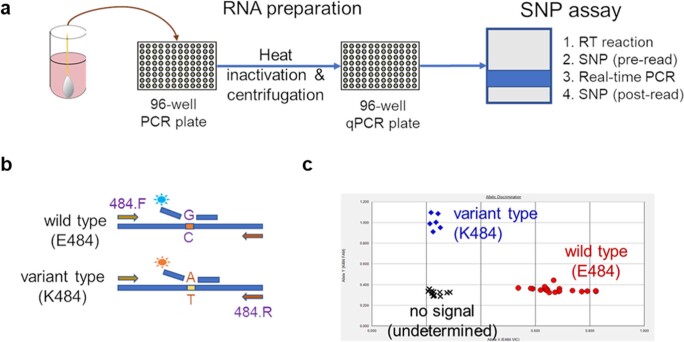
Extended Data Fig. 1. Rapid PCR-based screening assay protocol to identify samples harboring key substitutions.
a, Viral RNA is prepared by heat inactivation and centrifugation. The supernatant is then used for the SNP assay, which entails four steps: the reverse transcription (RT) reaction, pre-PCR reading of the plate to assess background fluorescence (SNP pre-read), real-time PCR, and post-PCR reading of the plate to measure fluorescence (SNP post-read). The runtime for this entire protocol is approximately two hours. b, Genotype at targeted sites in COVID-19 viral RNA can be determined with two MGB probes, one for wild type (conjugated with VIC) and the other for variant type (conjugated with FAM). c, Example signals for the variant type (K484; blue), the wild type (E484; red) and samples with no signal (black) are shown.