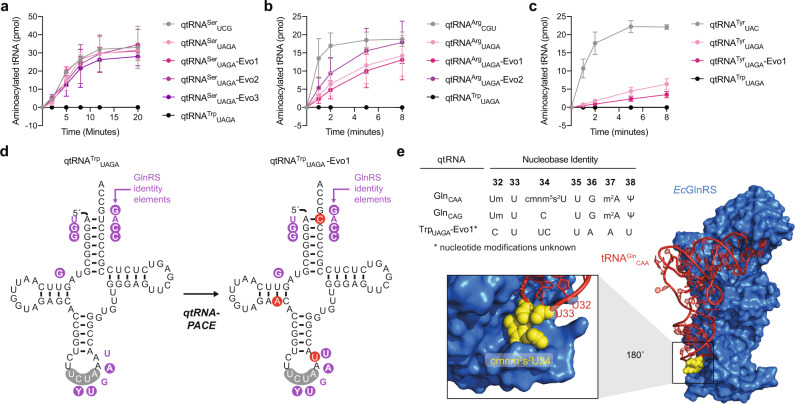
Fig. 3. In vitro analysis of evolved qtRNAs.
In vitro aminoacylation of qtRNAs using EcSerRS (a), EcArgRS (b), and EcTyrRS (c); qtRNATrpUAGA was used as a negative control in all cases. d Analysis of EcGlnRS identity elements as found in the engineered qtRNATrpUAGA and qtRNA-PACE evolved qtRNATrpUAGA-Evo1. e Anticodon and adjacent nucleobase identities for known E. coli glutamine tRNAs and the qtRNA-PACE evolved qtRNATrpUAGA-Evo1. Position of the modified nucleotide cmnm5s2U (yellow) in the co-crystal structure of tRNAGlnCAA and E. coli GlnRS (4JYZ; https://www.wwpdb.org/pdb?id=pdb_00004jyz). Data represent the mean and standard deviation of three biological replicates.