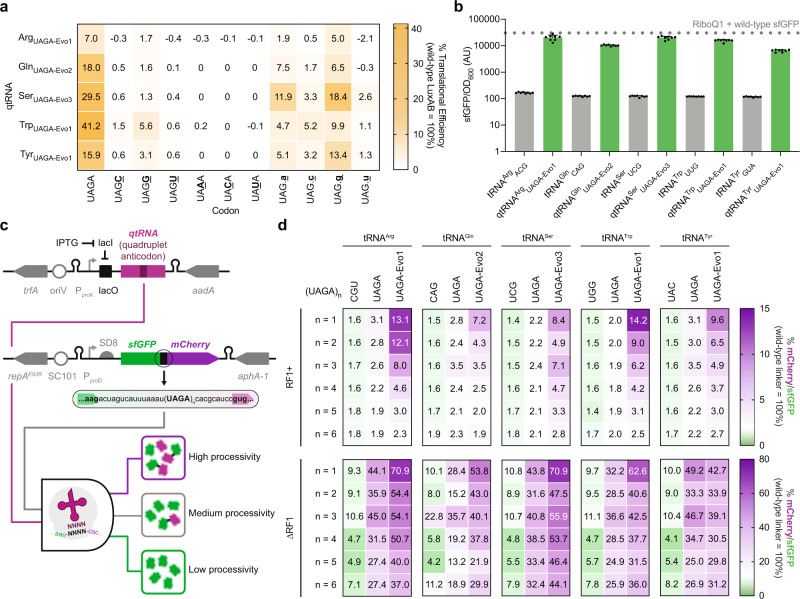
Fig. 4. Quantification of evolved qtRNA crosstalk and processivity.
a Evolved UAGA-qtRNAs show moderate crosstalk with codons bearing a different nucleotide at the fourth position but not the third position of the quadruplet codon. Evolved UAGA-qtRNAs can additionally crosstalk with amber (UAG) codons, leading to a frameshift that is dependent on the base following the stop codon. In all cases, the activity of non-cognate codon-anticodon pairs is lower than the expected cognate interactions. b Quadruplet-codon translation using a previously described orthogonal ribosome RiboQ1 (contains mutations U531G, U534A, A1196G, A1197G in the 16S rRNA)11 (n = 8 biologically independent samples). c Schematic representation of the dual sfGFP-mCherry reporter assay to investigate the processivity of quadruplet codon translation. In all cases, sfGFP and mCherry are separated by a linker composed of adjacent UAGA quadruplet codons. The ratio of mCherry to sfGFP is a proxy for UAGA codon suppression efficiency and processivity. d Evolved UAGA qtRNAs effectively translate linkers containing up to 5−6 adjacent UAGA quadruplet codons in the RF1 + strain S3489 (top) or the ∆RF1 strain JX33 (bottom). Although showing moderate efficiencies, these tandem suppression events have been validated using mass spectrometry (wherever possible) and comprehensive peptide fragmentation spectra are reported in Supplementary Fig. 10, and summary LC-MS/MS results are reported in Supplementary Table 8. In all cases, reporter data is normalized to an otherwise wild-type protein and color-coded by the used reporter. Data represent the mean and standard deviation as appropriate.