Abstract
Most organisms contain self-sustained circadian clocks. These clocks can be synchronized by environmental stimuli, but can also oscillate indefinitely in isolation. In mammals this is true at the molecular level for the majority of cell types that have been examined. A core set of “clock genes” form a transcriptional/translational feedback loop (TTFL) which repeats with a period of approximately 24 hours. The exact mechanism of the TTFL differs slightly in various cell types, but all involve similar family members of the core cohort of clock genes. The clock has many outputs which are unique for different tissues. Cells in diverse tissues will convert the timing signals provided by the TTFL into uniquely orchestrated transcriptional oscillations of many clock-controlled genes and cellular processes.
Keywords: Circadian rhythms, clock genes, chronobiology, biological rhythms
1.1. INTRODUCTION
One feature common to how organisms evolved on Earth was the presence of a 24-hour rhythm produced by the planet’s rotation about its axis. During a 24-hour period, several environmental changes occur including variations in light, temperature, humidity, and availability of food. Over millennia, these oscillations have created evolutionary pressures for organisms, both simple and complex, to develop physiological rhythms that allow them to anticipate these daily changes. Examples of mammalian behaviors that have a circadian basis include sleep/wake cycles, body temperature fluctuations, and circulating hormone levels [1–4]. Disruptions of the circadian rhythm may contribute to a number of human disorders and diseases ranging from Delayed and Advanced Sleep Phase Syndromes, jet lag, memory impairment, and shift work sleep disorder, to various other psychiatric, neurological, and metabolic diseases [5–8]. The impact of circadian dysfunction may in fact be greater than what is currently appreciated as diseases that do not have obvious connections to the circadian system may also be involved. Phenomena such as diabetes and hypoinsulinemia, myocardial infarction and asthma, susceptibility to certain cancers, and even the efficacy of certain medical treatments may all have a circadian component [9–12].
Work over the last several decades has revealed that our internal timekeeping ability is due to a series of molecular events that occur at the cellular level. The canonical model of the molecular clock involves a number of interlocking transcriptional-translational feedback loops with built-in delays that enable robust rhythms that last roughly 24-hours. These molecular clocks can synchronize or “entrain” to input from external stimuli so that the organism can adapt to the changing environment. But these clocks are also able to “free-run” which means to persist even in the absence of any external entraining stimuli. Other significant discoveries show that non-transcriptional modifications, such as post-transcriptional, translational, and post-translational changes, all play a significant role in the function and regulation of the molecular circadian clock. Finally, the outputs of the clock or clock controlled genes are unique among various tissues throughout the body. A discussion of the highlights of these developments will be provided in later sections.
1.2. The Molecular Circadian Clock in Mammals
1.2.1. Transcriptional Rhythms
The molecular circadian clock in mammals is a cell-autonomous process that is driven by a system of interlocking positive and negative transcriptional-translational feedback loops (TTFLs). The primary TTFL, or at least the first described, involves approximately 10 genes and includes the ‘core’ clock genes, Clock, Bmal1, Period (Per1, Per2, Per3), and Cryptochrome (Cry1, Cry2). Together, they function to produce robust rhythms of gene expression that repeats approximately every 24 hours (Figure 1).
FIGURE 1.
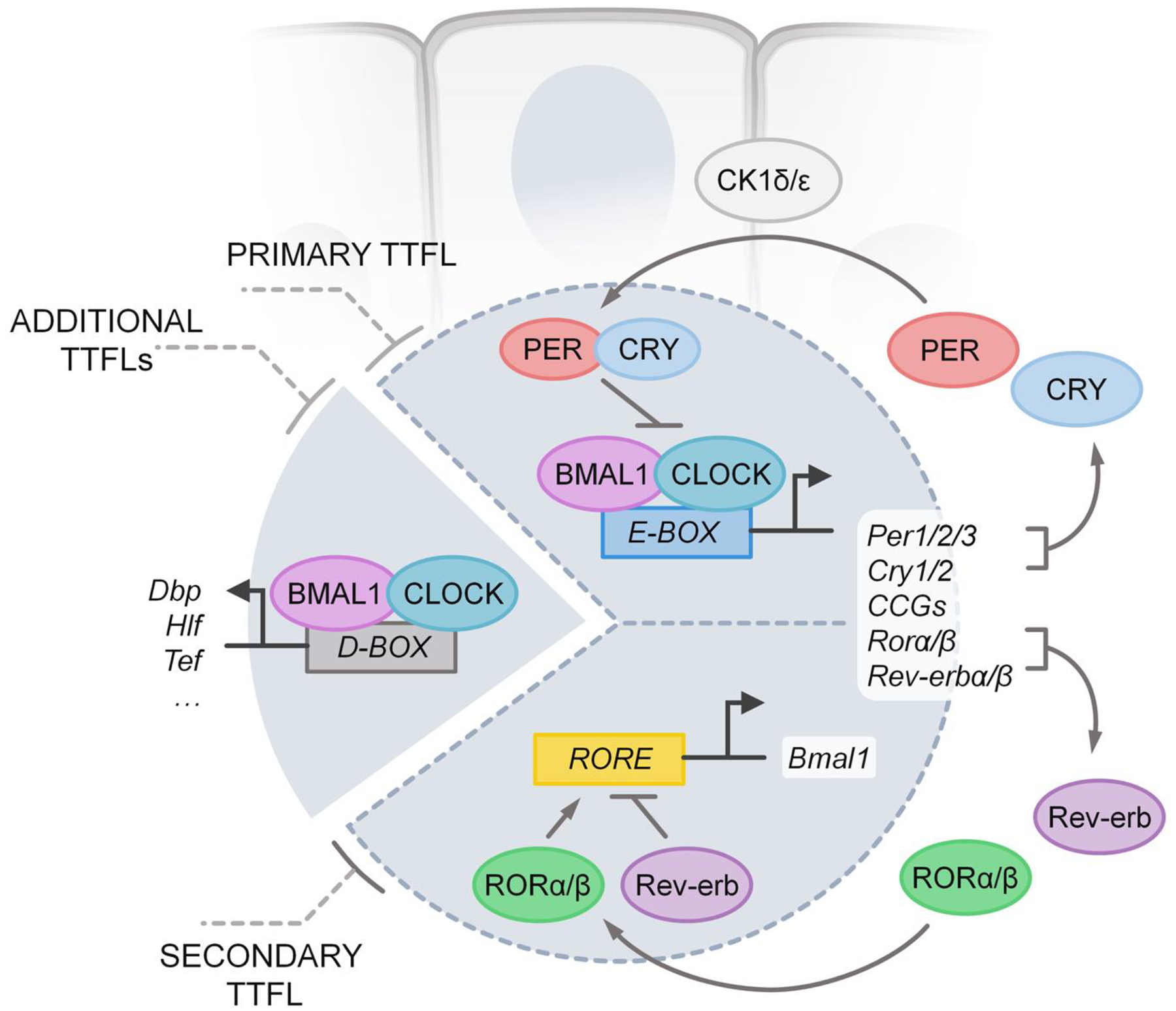
The core molecular circadian transcriptional/translational feedback loop in mammals.
The circadian clock starts in the morning, or early light phase, with the transcription of the positive limb of the main TTFL which is composed of the transcription-activating clock genes, Clock and Bmal1 [13–16]. Once translated, their gene products dimerize into the CLOCK:BMAL1 protein complex to form the heterodimeric basic HLH (helix-loop-helix)-PER-ARNT-SIM (bHLH-PAS) transcription factor. This CLOCK:BMAL1 complex then binds to the CACGTG E/E’-box DNA response elements found in the promoter or enhancer regions of target genes to promote their transcription [17–19]. In the mouse, peak accumulation of CLOCK:BMAL1 complex in the nucleus occurs in the dark phase [20]. The ability for the CLOCK:BMAL1 complex to bind many target genes is an important feature of the circadian clock because it represents a means by which oscillations of the core clock genes can drive transcription of not only the clock genes themselves, but also clock-controlled genes throughout the body. The temporally-controlled transcription of genes may be useful to not only regulate the abundance of given transcripts, but also to efficiently coordinate the transcriptional machinery itself [21].
One set of genes activated by the CLOCK:BMAL1 complex includes the negative limb of the main TTFL which is composed of the clock genes, Period (Per1, Per2, Per3) and Cryptochrome (Cry1, Cry2). PER and CRY proteins dimerize and accumulate outside of the nucleus with peak accumulation occurring in the late afternoon or evening [20]. After a time lag, the PER:CRY complexes translocate into the nucleus at night and act on the CLOCK:BMAL1 complex to inhibit its transcriptional activity [20, 22]. The mechanism of repression appears to involve the direct binding of the CRY proteins to the CLOCK:BMAL1 complex [23–26], and the shuttling of kinases such as Casein Kinase I delta (CK1δ) into the nucleus [27, 28]. However, the inhibitory role of the PER proteins is less clear. For example, studies have shown that even in the absence of PER proteins, CRY proteins can inhibit CLOCK:BMAL1-mediated transcription, binding directly to transactivation domains (TADs) on BMAL1 [24–26]. PER proteins, on the other hand, do not bind to the CLOCK:BMAL1:Ebox complex, but instead seem to promote CLOCK:BMAL1 dissociation from DNA binding elements in association with CRY proteins [23, 27, 29, 30]. Additional work will help to fully understand the exact mechanisms of the repressive proteins. For example, the protein complex including CRY:PER dimers has been found to be as large as 1.9 MDa inside the nucleus, containing many proteins in addition to the ones described here [31].
A secondary TTFL runs alongside the main TTFL. This secondary loop is initiated by the CLOCK:BMAL1 complex which promotes the E-box mediated transcription of the orphan nuclear receptor genes REV-ERBα/β and RORα/β [32–34]. Their protein products, REV-ERB and ROR, then compete for the retinoic acid-related orphan receptor response elements (RORE) binding sites within the promoter of Bmal1 [32–34]. The transcription of Bmal1 is inhibited when REV-ERB proteins are bound to the RORE binding sites, while the binding of ROR proteins promotes it [35]. Individual genetic knockouts of REV-ERBα, both in mice and in cultured cells, only showed mild to moderate period and rhythm phenotypes; however, when REV-ERBα and β were deleted in the same animal, a more severe loss of rhythmicity was observed [32, 35, 36]. Cistromic analysis of REV-ERB α/β and BMAL1 have shown that overlapping genes are highly enriched for clock and metabolic functions, including lipid metabolism [35]. Another potential TTFL that is initiated by the CLOCK:BMAL1 complex involves the transcription of the PAR bZIP genes that contain D-box elements in their promotors, such as D-box binding protein (DBP), hepatic leukemia factor (HLF), thyrotroph embryonic factor (TEF), and nuclear factor interleukin 3 (NFIL3) [37].
An additional member of the core clock gene family may have been discovered with the assistance of a machine learning approach to large datasets. The existence of the computationally highlighted repressor of the network oscillator (CHRONO) protein was first flagged as a candidate by sequence homology to other repressor genes. It was then further drawn out of a screen employing a two-hybrid analyses in mouse fibroblast cells that investigated the highest affinity binding targets of the CLOCK:BMAL1 complex [38, 39]. Subsequent studies have shown that CHRONO expression is antiphasic to Bmal1 and, similar to the CRY proteins, transcriptionally represses CLOCK:BMAL1. The mechanism of repression may involve histone modification by histone deacytlase (HDAC) [40]. Further, the PAS domain containing repressor 1, PASD1 has also been shown to repress the CLOCK:BMAL1 complex and dampen the oscillations of the molecular clock [41]. As the science progresses, it will be exciting to see how new discoveries will continue to change our understanding of the molecular circadian clock and of how it is regulated.
The chromatin remodeling necessary for the cyclic transcriptional activity of the main TTFL is achieved by the rhythmic acetylation and deacetylation of the H3 and H4 histones, and is facilitated by clock-specific and ubiquitous histone-modifying proteins. When the circadian clock first starts, the activators, CLOCK and BMAL1, interact with the histone acetyltransferases (HAT) p300 and CREB binding protein (CBP), respectively, in order to remodel chromatin to a state that is accessible for transcription [42–45]. The CLOCK protein itself has also been reported to have a HAT domain that acetylates the histones H3K9 and H3K14 [46, 47]. In addition, CLOCK:BMAL1 complex has been shown to recruit the methyltransferase MLL1 (mixed lineage leukemia 1) to cyclically methylated histone H3 and the histone deacetylase (HDAC) inhibitor, JARID1a, which further facilitates transcriptional activation [48, 49]. During the repressive phase of the circadian clock, deacetylation occurs, at least in part, due to the recruitment of PER1 and the SIN3-HDAC complex to CLOCK:BMAL1-bound DNA [50]. In addition, SIRT1, a NAD+ dependent HDAC, also associates with CLOCK, BMAL1, PER2, and PGC1α to ultimately inhibit the CLOCK:BMAL1 complex and decrease its transcriptional activity [51–55].
While much attention is given to the CLOCK:BMAL1 complex, Reppert and colleagues were surprised to find that Clock-deficient mice showed normal behavioral rhythmicity and functional, but reduced, TTFL molecular activity [56]. The same group then identified NPAS2 as a functionally redundant partner of BMAL1 in CLOCK’s absence [57]. Intriguingly, this CLOCK/NPAS2 redundancy is primarily observed in the brain’s master clock (see section 1.5), whereas most peripheral clocks throughout the body require the CLOCK protein itself as an obligate partner for BMAL1 [58].
Finally, it should be noted that the presence of transcriptional rhythm does not always correlate directly with translation/protein abundance rhythms, and vice versa. Studies have found that although 20% of soluble proteins of the mouse liver oscillate, only about half of those have a corresponding oscillatory transcript [75–77].
1.3. POST-TRANSCRIPTIONAL REGULATION OF THE CIRCADIAN CLOCK
Historically, the circadian field has focused on understanding the transcriptional mechanisms of the molecular clock. However, much work has provided insights into the important role that non-transcriptional processes play in setting the pace of the clock. Processes such as post-transcriptional, translational, and post-translational modifications have all been shown to be significant. A brief summary of recent discoveries will be provided here, although more thoughtful reviews can be found [59, 60].
1.3.1. Post-transcriptional modifications of RNA
Post-transcriptional modifications, such as that for mRNA, are ubiquitous in biology, and circadian biology is no exception. Although evidence suggests that the core clock genes generally appear to be rhythmically transcribed [21, 39, 61], this does not mean that all genes that are rhythmically transcribed are transcribed de novo. In the liver, for example, 10% of genes are rhythmic but only about 20% of those are expressed de novo and may be explained by mRNA processing [62]. There is also evidence that suggests post-transcriptional modifications can directly impact the period of the clock. One study examined the effects of selective N6-methylation of adenosine (m6A), which is present on many gene transcripts, including clock gene transcripts such as Period (Per1, Per2, Per3), Clock, Bmal1, Ck1δ, and Rev-Erb α. They found that inhibiting methylation lengthened the circadian period while promoting methylation shortened the it [63, 64]. The 3’ UTR of clock gene transcripts also appears to play a significant role in their stability and degradation as studies have shown that Per2 transcripts that either lacked their 3’ UTR or had the endogenous Per2’ 3’UTR substituted with an SV40 late poly(A) tail had a longer half-life than those without either modification [65, 66]. Alterations to the 3’ UTR of other clock gene transcripts including the Period genes, Cry1, and Bmal1 all appear to significantly affect the length of the circadian period [66–74].
1.3.2. Translational modifications
Translational regulation may account for the discrepancy between transcriptional and translational rhythmicity. One way this can occur is in a cell’s ability to vary its translational efficiency by changing the availability and activity of its ribosomes. This is supported by recent work from Janich et al. where they showed that, depending on the circadian phase, there was significant variation in ribosomal occupancy in the mouse liver for many gene transcripts, including clock genes like Clock, Bmal1, Cry1, and Rev-Erbα/β. They saw that, despite large variations in the quantity of clock gene transcripts, the quantity of their respective clock proteins were still comparable with one another [61, 78, 79]. The exact mechanism by which this occurs has yet to be fully elucidated but likely involves the presence of upstream open reading frames (uORFS) in the 5’UTR of these transcripts, though this may not always be the case [61, 80, 81]. Other translational regulators, such as mTOR, have also been shown to acutely affect the translation of circadian proteins such as PER and CRY in response to stimuli that normally influences the circadian clock, such as insulin and IGF-1 [72].
1.3.3. Post-translational protein modifications
In order for the circadian clock to restart a new cycle of transcription, the levels of PER and CRY must decrease sufficiently such that repression is relieved. How quickly this happens, however, depends largely on the stability of the repressive proteins. Unsurprisingly, variations in their stability and rate of degradation can affect the free-running period at which the clock runs. For example, the tau mutant hamster was the first mammal identified as a circadian mutant and exhibited a free-running period of about 20 hours compared to the 24-hour free-running period of their wild-type counterparts [82]. The mutant tau locus was identified as a hypomorphic allele of Casein Kinase I epsilon (CKIε) which is an enzyme that phosphorylates the PER proteins [83]. Biochemical work since this initial discovery has shown that the stability of the PER proteins is primarily regulated by two Casein Kinases (CKIε and CK1δ), as well as phosphatase 1 [28, 83–86]. For instance, CKIε/δ-mediated phosphorylation targets the PER proteins for ubiquitination by β–TrCP and subsequent degradation by the 26S proteasome, ultimately shortening the period of the circadian clock [87–93]. The ‘phosphoswitch’ model of PER2, which encapsulates our most current understanding of this mechanism, was first introduced in 2015 by Zhou et al [91]. According to this model, the stability of PER2 is regulated by kinase activity at two antagonistic regulatory regions that determine whether the degradation rate of PER2 is fast or slow. The first region, ‘FASP,’ resides at S659 on the PER2 protein and is located within the Casein Kinase binding domain (CKBD). FASP has five separate sites that, when phosphorylated, increases its stability and slows its rate of degradation. Phosphorylation is mediated by either CKIε, CKIδ or both [94] and is initiated sequentially, starting with the first non-consensus serine. The second region termed ‘phosphodegron’ is located after the PAS-B domain at S478 on PER2 which is several hundred residues upstream of the CKBD. Ultimately, PER2 stability appears to be due to the balance between the phosphorylation states of these two antagonistic regions.
Despite this model’s explanatory power, the regulatory roles that kinases have on PER2 may be more complex. For example, a recent study examined a PER2-Ser478Ala knock-in mutant mouse line. They found that behavioral rhythms were statistically different and increased by about an hour, comparable to what was observed in mice who were injected with a CKI inhibitor [95, 96]. Interestingly, however, the difference in period length of PER2-Ser478Ala mutant MEFs versus their controls does not seem to be consistent with what would be expected from such a mutation, if any statistically significant difference can be found at all [96]. These findings suggest that there are compensatory mechanisms affecting PER2 stability that do not require the phosphodegron at S478. Casein Kinase 2 (CK2) also phosphorylates PER2 and promotes its degradation via pathways that can be CKIε/δ-independent [97–99]. A more complicated model may also be required for other Period paralogs. PER1 does not have a ‘phosphodegron’ at S478, though it does possess a different phosphodegron motif that, when phosphorylated, still recruits β–TrCP to initiate its ubiquitination and subsequent degradation [89, 92]. PER1, like PER2, also has at least one kinase other than CKIε/δ that can regulate its stability [100].
Experiments have also shown that the stability and degradation rate of CRY proteins impacts the period of the circadian clock. Two studies analyzing chemically-induced mutations in mice found that mutations in the F-box gene, Fbxl3, resulted in longer periods of behavioral rhythms [101, 102]. FBXL3 polyubiquitinates CRY proteins, marking them for proteasomal degradation in the SKP1-CUL1-F-box protein E3 ubiquitin ligase pathway (SCF) [101–103]. Interestingly, the CRY proteins act as cofactors with the SCFFbxl3 complex for at least 100 protein targets, some likely to be clock regulated and some not [104]. FBXL21, the closest paralog of FBXL3, ubiquitinates and stabilizes the CRY proteins, attenuating the period-lengthening effects of FBXL3 knockouts [105, 106]. In terms of CRY degradation, the pathways differ between the CRY paralogs as CRY1 is phosphorylated by AMPK1 while CRY2 by the DYRK1A/GSK-3B cascade [107–109]. Once PER and CRY levels become sufficiently low as the night progresses, the repressive phase ceases and the TTFL reinitiates.
Significant work has been done to identify kinases acting on other clock proteins such as CLOCK, BMAL1, REV-ERBα and ROR. GSK3B, for example, has been shown to phosphorylate CLOCK, BMAL1 and CRY 2 and promote proteasomal degradation, while its phosphorylation of REV-ERBα promotes its stability [109–112]. Intriguingly, GSK3B has also been shown to impact length of the circadian period by promoting nuclear localization of PER2 (CKIε and CKδ appears to regulate the subcellular localization of PER1 and PER3) [113–115]. Other examples include CDK5 which phosphorylates CLOCK and increases its stability and transcriptional activity, dual-specificity tyrosine-phosphorylated and regulated kinase 1A (DYRK1A) and GSK3B which phosphorylates CRY2 and promotes its degradation, AMP-activated protein kinase (AMPK) which phosphorylates CRY1 and promotes its degradation, and RORα, which is negatively regulated by extracellular signal-related kinase 2 (ERK2)-mediated phosphorylation. Given the role that kinases have in the post-translational modification of clock proteins, it is unsurprising that phosphatases are also important. Protein phosphatase 1 (PP1) activity, for example, promotes PER2 stabilization while protein phosphatase 5 (PP5) appears to increase circadian period length and decrease PER expression in fibroblasts, in vitro [86, 116, 117]. These modifications can impact not just their stability, but their transcriptional activity, nuclear localization, and interaction with other proteins, as well.
Other post-translational modifications are also significant in the regulation of the circadian clock. The sumolyation of BMAL1 by SUMO2/3, for example, is rhythmic and promotes both its transcriptional activity and degradation while mutation at one sumolyation site, K259R, lengthens its half-life [44, 118]. BMAL1 is also acetylated by histone acetyltransferase, TIP60, which causes elongation of transcripts that are targeted by the CLOCK:BMAL1 complex [119]. There is also evidence that suggests that core clock proteins such as CLOCK and PER undergo O-GlcNAcylation and it is hypothesized that this process serves as a metabolic sensor for the cell, working intricately with kinases to regulate the circadian clock [120]. Altogether, these insights provide a glimpse into the immense importance that non-transcriptional processes have in the regulation of the circadian clock and will be an undoubtedly be an exciting area to watch as the research advances.
1.4. CLOCK-CONTROLLED GENOME-WIDE TRANSCRIPTION (CLOCK OUTPUTS)
A fundamentally important feature of the molecular circadian clock is that it generates ‘clock outputs’ as it oscillates throughout the day. These clock outputs are the means by which the oscillations of the core clock genes drive transcriptional programs at specific times in various tissues throughout the body. The main clock outputs are driven by CLOCK:BMAL1, REV-ERB/ROR and DBP, and they can affect not just the clock genes but thousands of other non-clock genes as well [60, 121, 122]. Recent advances in genomic technologies such as chromatin immunoprecipitation with sequencing (ChIP-seq) have improved our understanding of the circadian cistrome, elucidating DNA binding sites of multiple core clock genes [21, 39, 123–125]. For example, nearly 300 genes are rhythmically expressed in the retina in constant darkness, but that number skyrockets to almost 3000 in the presence of light [126]. Other studies have shown that between 2 and 10% of all genes in select tissues of the mouse were expressed with a near 24-hour rhythm even though they were not considered to among the core clock genes [21, 62, 127–129]. A more recent examination of the transcriptomes of 12 mouse organs using RNA sequencing and DNA arrays found that 43% of all protein-coding gene transcripts in the mouse genome oscillated in at least one organ [130]. Unsurprisingly, these outputs can affect a variety of biological processes that, at first glance, may not be obviously tied to the rhythmic oscillations of the core clock genes. Work over the last two decades have revealed that gene families ranging from timing of the cell cycle to immune function to glucose and fatty acid metabolism appear to be intricately tied to these clock outputs [21, 62, 127–129, 131–133]. Intriguingly, even processes such as the recruitment of RNA polymerase, and large changes in chromosomal organization and chromatin topology have also been shown to be rhythmic in nature [60, 123, 124, 134–139].
Genes that are transcribed with a 24-hour rhythm vary greatly depending on the tissue. In fact, multiple studies have shown that there is a high degree of tissue-specificity in the genes that are expressed with minimal overlap between tissues. For example, one study that examined the transcriptional profiles between the SCN and liver in the mouse found that 337 genes were found to be rhythmically transcribed in the SCN compared to 335 genes in the liver, and only 28 genes overlapped [128]. Similarly, almost 450 oscillating genes were found to be expressed in the liver and heart, only 37 of which were in common [129]. For example, genes that are rhythmically expressed in the retina are involved in photoreception, synaptic transmission, and cellular metabolism, and this appears to be required for proper retina development [126]. The loss of core clock genes such as Bmal1 not only results in the loss of transcriptional oscillations, but compromised the functional integrity of the visual circuitry, likely by altered cell cycle kinetics during retinal neurogenesis [126, 140].
Transcriptional profiling has also ventured outside of the mouse model with a heroic collection of 64 tissues from baboons in a light:dark cycle over the course of a day by Mure, et al. [141]. Similar to the mouse transcriptomes, it was confirmed that all tissues contained extensive rhythmic gene expression profiles, and there was again surprisingly little overlap between tissues. They observed that 80% of all protein coding transcripts displayed rhythmic abundance in at least one tissue [141].
1.5. SCN AS THE MAIN SYNCHRONIZER AND THE ENTRAINMENT OF PERIPHERAL CLOCKS
A series of landmark experiments in the latter half of the 20th century changed the way we viewed circadian timekeeping in mammals. In 1972, Stephan and Zucker found that rhythmic drinking and locomotor activity in rats was lost when the suprachiasmatic nucleus (SCN) was lesioned [142], and Moore and Eichler found that similar lesions halted circulating corticosterone rhythms [143]. By 1990, Ralph and Menaker found that transplanting SCN from tau mutant hamsters onto SCN-lesioned and arrhythmic wild-type hosts not only restored rhythmic behavior but did so with shortened circadian periods [144]. These experiments, along with others, provided evidence that the SCN was sufficient for behavioral circadian timekeeping in mammals [142–145]. This gave rise to the hierarchical single-pacemaker model of the circadian system where the SCN, a small paired structure in the ventral hypothalamus containing as few as 10,000 neurons, was the ‘master’ clock that unilaterally controlled all rhythmic behavior throughout the body (Figure2) [146, 147].
Figure 2.
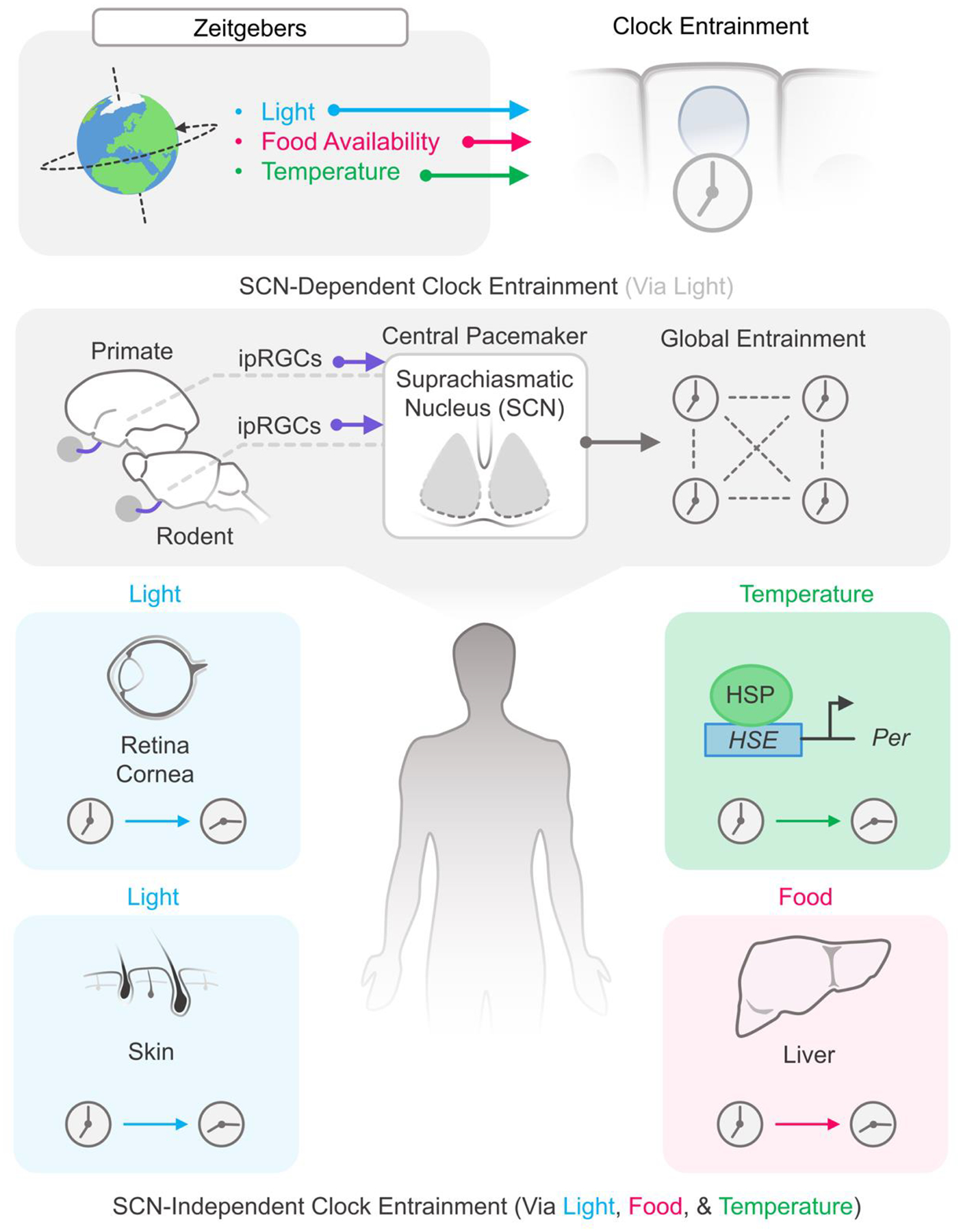
The hierarchy of temporal organization in mammals. Light cycles entrain the SCN, which in turn sets the rhythms for the body. In some instances, peripheral tissues can dissociate from typical central time using rhythmic environmental cues.
The earliest evidence suggesting that the single-pacemaker model may not be complete was actually observed in the 1950s—well before the SCN was ever thought to play that role. In these first experiments, the cultured intestines of golden hamsters were observed to have 24-hour rhythms of peristalsis, a behavior that could not have been explained by the rhythmicity of the SCN [148]. In the decades that followed, numerous other studies identified the circadian basis of other peripheral tissues throughout the body, including the heart, liver, and adrenal glands [149–154]. By the 1990s, molecular evidence for the presence of circadian clocks in mammalian peripheral tissues was established after the discovery of two mammalian core clock genes, Clock and Period [14, 15, 155, 156]. Since then, it has been shown that nearly all mammalian tissues have functional, cell-autonomous clocks with the same TTFLs that are described in the prior sections [157–159]. These studies showed that the fundamental unit of circadian time is kept not by the SCN but by the cell itself.
However, the discovery that almost all mammalian cells have functional and autonomous molecular clocks does not mean the SCN no longer plays a central role in circadian biology. In fact, even though the SCN does not control the molecular clocks of individual cells, it is still able to orchestrate the rhythms of diverse range of rhythmic behavior, from the sleep-wake cycle to feeding and fasting, the regulation of temperature, and the circulation of hormones throughout the body [1–4]. The method by which the SCN is able to synchronize the innumerable clocks throughout the body occurs by behavioral, neuroendocrine, or autonomic signals [160–163].
Given its fundamental role in synchronizing tissues throughout the body, the SCN is still considered to be the principle pacemaker of the body. But a biological clock is only useful if it can be synchronized to an environmental signal and the SCN is no exception. In fact, the SCN itself can be entrained by light received by the retina, a necessary feature likely risen out of our evolutionary need to adapt to the 24-hour solar cycle. Although light information from rods, cones, and melanopsin-containing intrinsically photosensitive retinal ganglion cells (ipRGCs) are all sent to the SCN, only light information from melanopsin-containing ipRGCs are needed to set the circadian phase of the SCN [164–166]. The mechanism by which this occurs involves the modulation of the spontaneous firing rate (SFR) of the SCN [4]. When light information is received by the melanopsin-containing ipRGCs, neural signals travel towards the SCN via the retinohypothalamic tract (RHT). The RHT then releases the neuropeptides pituitary adenylate cyclase-activating polypeptide (PACAP) and glutamate within the core of the SCN [167]. These core SCN neurons propagate this signal to the rest of the SCN which alters its SFR and ultimately causes a rephasing of TTFLs in cells throughout the body [168]. Entrainment to light is also reflected in changes in transcription within the SCN—light causes an induction of genes in the Per family, the magnitude of which is dependent on the circadian time [169–171].
The single-pacemaker model where peripheral tissues only respond to signals from the SCN also appears to be outdated. A number of studies have shown that peripheral tissues themselves can become directly entrained by responding to environmental signals without the need for an SCN, such as light, temperature, and feeding. Though the signals by which different peripheral tissue entrain may vary, one environmental stimuli that may be universal across all peripheral tissues and cells is changes in body temperature. The 24-hour rhythmic component to body temperature fluctuations is mediated by the SCN, but the SCN itself is resistant to the changes [172–174]. The exact mechanism by which temperature entrainment occurs is still unclear but appears to involve Heat Shock Factors (HSF) and cold-induced RNA binding proteins (Cirbp). HSF are a family of DNA binding proteins that, when bound to Heat Shock Elements (HSE) located on the promoters of their target genes, lead to the translation of a number of proteins including those for Heat Shock Proteins (HSP). These HSPs facilitate the Heat Shock response, an essential pathway the cell utilizes to protect against protein-damaging stress. With respect to circadian biology, one study found that HSF binds rhythmically to HSEs and that the promoter region of Per2 contains HSEs. Interestingly, they also found that the expression of HSPs appear to oscillate in similar phase to the PERs and that PER2 levels transiently change in response to a heat pulse [175]. Another study which inhibited the HSF pathway with the compound KNK437 observed that responses to heat pulses were blocked and that the period of the circadian clock slowed to as long as 30 hours, which is consistent with what has been reported in HSF-1 deficient mice [173, 176]. The heat-mediated responses of Period gene expression and HSF1-mediated gene expression seem linked in that mutations to the HSE in the promoter of the Period gene dramatically altered its rhythm and heat response [177]. Cirbp acts in a unique way, in that it controls RNA stability of directly bound transcripts. Cirbp itself is not regulated as a clock output gene, but rather has activity which reflects the temperature experienced by the cell [178]. Interestingly, reduced temperatures allow for the splicing of Cirbp pre-mRNA into final mRNA for nascent protein to be produced and exert its effect on other RNA strands [179].
Since the discovery of peripheral clocks and their ability to be entrained by the SCN, there has, in parallel, been growing interest in non-SCN mediated entrainment of peripheral tissues. Restricted feeding cycles, particularly paradigms in which food is only available at atypical feeding times, cause the phase of the circadian clocks of the liver to alter their phase [180]. Remarkably, the phase of the liver will re-entrain to a new phase of food availability in an animal in which the SCN remains entrained to the environmental light cycle [181]. Even light itself may communicate with circadian clocks outside of the SCN pathway. In mice in which the molecular clock was only functional in the skin, and not the SCN or any other tissue, light cycles were still able to entrain the skin circadian clocks [182]. The circadian clocks of murine skin ex vivo were also found to be directly photoentrainable [183]. The area of skin and wavelength of light may be important for this effect [184, 185]. Interestingly, the process requires a subset of cells that express neuropsin (Opn5), a noncanonical opsin. These Opn5-expressing cells are expressed in dermal melanocytes and, when pulsed with a short-wavelength light (415nm), strongly shifts the circadian phase of murine skin ex vivo [183]. It was also observed that direct photoentrainment of circadian clocks in the retina and cornea occurs in an Opn5-dependent manner [186, 187]. As the field moves forward, it will be exciting to see how new discoveries will elucidate the pathways by which external stimuli can directly entrain the molecular clocks of peripheral tissues and how they impact their underlying physiology.
1.6. GENERAL CONCLUSIONS
The field of circadian biology has exploded from observations of the leaf movements of potted plants in the 1700’s [188] to modeling and observing individual protein motifs involved in circadian molecular dynamics today. From the initial discoveries that a single heritable mutation could change the circadian rhythm of fly behavior [189–191], to the remarkable detail of the molecular clockwork mechanism described above, the field has expanded to influence many diverse areas of physiology. A current push is underway to apply these amazing findings to the benefit of human health. The term “chronotherapy” will likely become commonplace as the timing of procedures and drug administration will soon be determined based on the state of the individual’s molecular clock [192].
Acknowledgements:
This work was supported by NIH R01 GM124246 to EDB, the Latham Vision Research Innovation Award, and an unrestricted grant to the University of Washington Department of Ophthalmology from Research to Prevent Blindness. We thank Richard A. Lang for his support of S.D’S.
Footnotes
Publisher's Disclaimer: This is a PDF file of an unedited manuscript that has been accepted for publication. As a service to our customers we are providing this early version of the manuscript. The manuscript will undergo copyediting, typesetting, and review of the resulting proof before it is published in its final form. Please note that during the production process errors may be discovered which could affect the content, and all legal disclaimers that apply to the journal pertain.
REFERENCES
- 1.Brown SA and Azzi A, Peripheral circadian oscillators in mammals. Handb Exp Pharmacol, 2013(217): p. 45–66. [DOI] [PubMed] [Google Scholar]
- 2.Czeisler CA, SLEEP. Measuring the passage of brain time. Science, 2016. 353(6300): p. 648–9. [DOI] [PubMed] [Google Scholar]
- 3.Potter GD, et al. , Circadian Rhythm and Sleep Disruption: Causes, Metabolic Consequences, and Countermeasures. Endocr Rev, 2016. 37(6): p. 584–608. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Hastings MH, Maywood ES, and Brancaccio M, Generation of circadian rhythms in the suprachiasmatic nucleus. Nat Rev Neurosci, 2018. 19(8): p. 453–469. [DOI] [PubMed] [Google Scholar]
- 5.Frank E, et al. , Influencing circadian and sleep-wake regulation for prevention and intervention in mood and anxiety disorders: what makes a good homeostat? Ann N Y Acad Sci, 2014. 1334: p. 1–25. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Hastings MH and Goedert M, Circadian clocks and neurodegenerative diseases: time to aggregate? Curr Opin Neurobiol, 2013. 23(5): p. 880–7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Panda S, Circadian physiology of metabolism. Science, 2016. 354(6315): p. 1008–1015. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Smarr BL, et al. , A time to remember: the role of circadian clocks in learning and memory. Behav Neurosci, 2014. 128(3): p. 283–303. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Muller JE, et al. , Circadian variation in the frequency of onset of acute myocardial infarction. N Engl J Med, 1985. 313(21): p. 1315–22. [DOI] [PubMed] [Google Scholar]
- 10.Stephenson R, Circadian rhythms and sleep-related breathing disorders. Sleep Med, 2007. 8(6): p. 681–7. [DOI] [PubMed] [Google Scholar]
- 11.Gaddameedhi S, et al. , Control of skin cancer by the circadian rhythm. Proc Natl Acad Sci U S A, 2011. 108(46): p. 18790–5. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Cederroth CR, et al. , Medicine in the Fourth Dimension. Cell Metab, 2019. 30(2): p. 238–250. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Vitaterna MH, et al. , Mutagenesis and mapping of a mouse gene, Clock, essential for circadian behavior. Science, 1994. 264(5159): p. 719–25. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Antoch MP, et al. , Functional identification of the mouse circadian Clock gene by transgenic BAC rescue. Cell, 1997. 89(4): p. 655–67. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.King DP, et al. , Positional cloning of the mouse circadian clock gene. Cell, 1997. 89(4): p. 641–53. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Bunger MK, et al. , Mop3 is an essential component of the master circadian pacemaker in mammals. Cell, 2000. 103(7): p. 1009–17. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Gekakis N, et al. , Role of the CLOCK protein in the mammalian circadian mechanism. Science, 1998. 280(5369): p. 1564–9. [DOI] [PubMed] [Google Scholar]
- 18.Hogenesch JB, et al. , The basic-helix-loop-helix-PAS orphan MOP3 forms transcriptionally active complexes with circadian and hypoxia factors. Proc Natl Acad Sci U S A, 1998. 95(10): p. 5474–9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Huang N, et al. , Crystal structure of the heterodimeric CLOCK:BMAL1 transcriptional activator complex. Science, 2012. 337(6091): p. 189–94. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Lee C, et al. , Posttranslational mechanisms regulate the mammalian circadian clock. Cell, 2001. 107(7): p. 855–67. [DOI] [PubMed] [Google Scholar]
- 21.Koike N, et al. , Transcriptional architecture and chromatin landscape of the core circadian clock in mammals. Science, 2012. 338(6105): p. 349–54. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Lowrey PL and Takahashi JS, Genetics of circadian rhythms in Mammalian model organisms. Adv Genet, 2011. 74: p. 175–230. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Ye R, et al. , Biochemical analysis of the canonical model for the mammalian circadian clock. J Biol Chem, 2011. 286(29): p. 25891–902. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Kondratov RV, et al. , Dual role of the CLOCK/BMAL1 circadian complex in transcriptional regulation. FASEB J, 2006. 20(3): p. 530–2. [DOI] [PubMed] [Google Scholar]
- 25.Shearman LP, et al. , Interacting molecular loops in the mammalian circadian clock. Science, 2000. 288(5468): p. 1013–9. [DOI] [PubMed] [Google Scholar]
- 26.Xu H, et al. , Cryptochrome 1 regulates the circadian clock through dynamic interactions with the BMAL1 C terminus. Nat Struct Mol Biol, 2015. 22(6): p. 476–484. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Cao X, et al. , Molecular mechanism of the repressive phase of the mammalian circadian clock. Proc Natl Acad Sci U S A, 2021. 118(2). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Etchegaray JP, et al. , Casein kinase 1 delta regulates the pace of the mammalian circadian clock. Mol Cell Biol, 2009. 29(14): p. 3853–66. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Ye R, et al. , Dual modes of CLOCK:BMAL1 inhibition mediated by Cryptochrome and Period proteins in the mammalian circadian clock. Genes Dev, 2014. 28(18): p. 1989–98. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Chiou YY, et al. , Mammalian Period represses and de-represses transcription by displacing CLOCK-BMAL1 from promoters in a Cryptochrome-dependent manner. Proc Natl Acad Sci U S A, 2016. 113(41): p. E6072–E6079. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Aryal RP, et al. , Macromolecular Assemblies of the Mammalian Circadian Clock. Mol Cell, 2017. 67(5): p. 770–782.e6. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Preitner N, et al. , The orphan nuclear receptor REV-ERBalpha controls circadian transcription within the positive limb of the mammalian circadian oscillator. Cell, 2002. 110(2): p. 251–60. [DOI] [PubMed] [Google Scholar]
- 33.Sato TK, et al. , A functional genomics strategy reveals Rora as a component of the mammalian circadian clock. Neuron, 2004. 43(4): p. 527–37. [DOI] [PubMed] [Google Scholar]
- 34.Guillaumond F, et al. , Differential control of Bmal1 circadian transcription by REV-ERB and ROR nuclear receptors. J Biol Rhythms, 2005. 20(5): p. 391–403. [DOI] [PubMed] [Google Scholar]
- 35.Cho H, et al. , Regulation of circadian behaviour and metabolism by REV-ERB-alpha and REV-ERB-beta. Nature, 2012. 485(7396): p. 123–7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Liu AC, et al. , Redundant function of REV-ERBalpha and beta and non-essential role for Bmal1 cycling in transcriptional regulation of intracellular circadian rhythms. PLoS Genet, 2008. 4(2): p. e1000023. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Yoshitane H, et al. , Functional D-box sequences reset the circadian clock and drive mRNA rhythms. Commun Biol, 2019. 2: p. 300. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Hatanaka F, et al. , Genome-wide profiling of the core clock protein BMAL1 targets reveals a strict relationship with metabolism. Mol Cell Biol, 2010. 30(24): p. 5636–48. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Rey G, et al. , Genome-wide and phase-specific DNA-binding rhythms of BMAL1 control circadian output functions in mouse liver. PLoS Biol, 2011. 9(2): p. e1000595. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Goriki A, et al. , A novel protein, CHRONO, functions as a core component of the mammalian circadian clock. PLoS Biol, 2014. 12(4): p. e1001839. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Michael AK, et al. , Cancer/Testis Antigen PASD1 Silences the Circadian Clock. Mol Cell, 2015. 58(5): p. 743–54. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Etchegaray JP, et al. , Rhythmic histone acetylation underlies transcription in the mammalian circadian clock. Nature, 2003. 421(6919): p. 177–82. [DOI] [PubMed] [Google Scholar]
- 43.Curtis AM, et al. , Histone acetyltransferase-dependent chromatin remodeling and the vascular clock. J Biol Chem, 2004. 279(8): p. 7091–7. [DOI] [PubMed] [Google Scholar]
- 44.Lee J, et al. , Dual modification of BMAL1 by SUMO2/3 and ubiquitin promotes circadian activation of the CLOCK/BMAL1 complex. Mol Cell Biol, 2008. 28(19): p. 6056–65. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Hosoda H, et al. , CBP/p300 is a cell type-specific modulator of CLOCK/BMAL1-mediated transcription. Mol Brain, 2009. 2: p. 34. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Doi M, Hirayama J, and Sassone-Corsi P, Circadian regulator CLOCK is a histone acetyltransferase. Cell, 2006. 125(3): p. 497–508. [DOI] [PubMed] [Google Scholar]
- 47.Hirayama J, et al. , CLOCK-mediated acetylation of BMAL1 controls circadian function. Nature, 2007. 450(7172): p. 1086–90. [DOI] [PubMed] [Google Scholar]
- 48.Katada S and Sassone-Corsi P, The histone methyltransferase MLL1 permits the oscillation of circadian gene expression. Nat Struct Mol Biol, 2010. 17(12): p. 1414–21. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.DiTacchio L, et al. , Histone lysine demethylase JARID1a activates CLOCK-BMAL1 and influences the circadian clock. Science, 2011. 333(6051): p. 1881–5. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Duong HA, et al. , A molecular mechanism for circadian clock negative feedback. Science, 2011. 332(6036): p. 1436–9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Nakahata Y, et al. , The NAD+-dependent deacetylase SIRT1 modulates CLOCK-mediated chromatin remodeling and circadian control. Cell, 2008. 134(2): p. 329–40. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Asher G, et al. , SIRT1 regulates circadian clock gene expression through PER2 deacetylation. Cell, 2008. 134(2): p. 317–28. [DOI] [PubMed] [Google Scholar]
- 53.Nakahata Y, et al. , Circadian control of the NAD+ salvage pathway by CLOCK-SIRT1. Science, 2009. 324(5927): p. 654–7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.Ramsey KM, et al. , Circadian clock feedback cycle through NAMPT-mediated NAD+ biosynthesis. Science, 2009. 324(5927): p. 651–4. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55.Foteinou PT, et al. , Computational and experimental insights into the circadian effects of SIRT1. Proc Natl Acad Sci U S A, 2018. 115(45): p. 11643–11648. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56.Debruyne JP, et al. , A clock shock: mouse CLOCK is not required for circadian oscillator function. Neuron, 2006. 50(3): p. 465–77. [DOI] [PubMed] [Google Scholar]
- 57.DeBruyne JP, Weaver DR, and Reppert SM, CLOCK and NPAS2 have overlapping roles in the suprachiasmatic circadian clock. Nat Neurosci, 2007. 10(5): p. 543–5. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 58.DeBruyne JP, Weaver DR, and Reppert SM, Peripheral circadian oscillators require CLOCK. Curr Biol, 2007. 17(14): p. R538–9. [DOI] [PubMed] [Google Scholar]
- 59.Crosby P and Partch CL, New insights into non-transcriptional regulation of mammalian core clock proteins. J Cell Sci, 2020. 133(18). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 60.Takahashi JS, Transcriptional architecture of the mammalian circadian clock. Nat Rev Genet, 2017. 18(3): p. 164–179. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 61.Janich P, et al. , Ribosome profiling reveals the rhythmic liver translatome and circadian clock regulation by upstream open reading frames. Genome Res, 2015. 25(12): p. 1848–59. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 62.Akhtar RA, et al. , Circadian cycling of the mouse liver transcriptome, as revealed by cDNA microarray, is driven by the suprachiasmatic nucleus. Curr Biol, 2002. 12(7): p. 540–50. [DOI] [PubMed] [Google Scholar]
- 63.Fustin JM, et al. , RNA-methylation-dependent RNA processing controls the speed of the circadian clock. Cell, 2013. 155(4): p. 793–806. [DOI] [PubMed] [Google Scholar]
- 64.Fustin JM, et al. , Two Ck1delta transcripts regulated by m6A methylation code for two antagonistic kinases in the control of the circadian clock. Proc Natl Acad Sci U S A, 2018. 115(23): p. 5980–5985. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 65.Woo KC, et al. , Mouse period 2 mRNA circadian oscillation is modulated by PTB-mediated rhythmic mRNA degradation. Nucleic Acids Res, 2009. 37(1): p. 26–37. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 66.Yoo SH, et al. , Period2 3’-UTR and microRNA-24 regulate circadian rhythms by repressing PERIOD2 protein accumulation. Proc Natl Acad Sci U S A, 2017. 114(42): p. E8855–E8864. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 67.Lee KH, et al. , AUF1 contributes to Cryptochrome1 mRNA degradation and rhythmic translation. Nucleic Acids Res, 2014. 42(6): p. 3590–606. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 68.Kim DY, et al. , hnRNP Q mediates a phase-dependent translation-coupled mRNA decay of mouse Period3. Nucleic Acids Res, 2011. 39(20): p. 8901–14. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 69.Kim SH, et al. , Rhythmic control of mRNA stability modulates circadian amplitude of mouse Period3 mRNA. J Neurochem, 2015. 132(6): p. 642–56. [DOI] [PubMed] [Google Scholar]
- 70.Nagel R, Clijsters L, and Agami R, The miRNA-192/194 cluster regulates the Period gene family and the circadian clock. FEBS J, 2009. 276(19): p. 5447–55. [DOI] [PubMed] [Google Scholar]
- 71.Chen R, D’Alessandro M, and Lee C, miRNAs are required for generating a time delay critical for the circadian oscillator. Curr Biol, 2013. 23(20): p. 1959–68. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 72.Crosby P, et al. , Insulin/IGF-1 Drives PERIOD Synthesis to Entrain Circadian Rhythms with Feeding Time. Cell, 2019. 177(4): p. 896–909 e20. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 73.Curtis AM, et al. , Circadian control of innate immunity in macrophages by miR-155 targeting Bmal1. Proc Natl Acad Sci U S A, 2015. 112(23): p. 7231–6. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 74.Lee KH, et al. , MicroRNA-185 oscillation controls circadian amplitude of mouse Cryptochrome 1 via translational regulation. Mol Biol Cell, 2013. 24(14): p. 2248–55. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 75.Mauvoisin D, et al. , Circadian clock-dependent and -independent rhythmic proteomes implement distinct diurnal functions in mouse liver. Proc Natl Acad Sci U S A, 2014. 111(1): p. 167–72. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 76.Reddy AB, et al. , Circadian orchestration of the hepatic proteome. Curr Biol, 2006. 16(11): p. 1107–15. [DOI] [PubMed] [Google Scholar]
- 77.Robles MS, Cox J, and Mann M, In-vivo quantitative proteomics reveals a key contribution of post-transcriptional mechanisms to the circadian regulation of liver metabolism. PLoS Genet, 2014. 10(1): p. e1004047. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 78.Jouffe C, et al. , The circadian clock coordinates ribosome biogenesis. PLoS Biol, 2013. 11(1): p. e1001455. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 79.Sinturel F, et al. , Diurnal Oscillations in Liver Mass and Cell Size Accompany Ribosome Assembly Cycles. Cell, 2017. 169(4): p. 651–663 e14. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 80.Atger F, et al. , Circadian and feeding rhythms differentially affect rhythmic mRNA transcription and translation in mouse liver. Proc Natl Acad Sci U S A, 2015. 112(47): p. E6579–88. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 81.Jang C, et al. , Ribosome profiling reveals an important role for translational control in circadian gene expression. Genome Res, 2015. 25(12): p. 1836–47. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 82.Ralph MR and Menaker M, A mutation of the circadian system in golden hamsters. Science, 1988. 241(4870): p. 1225–7. [DOI] [PubMed] [Google Scholar]
- 83.Lowrey PL, et al. , Positional syntenic cloning and functional characterization of the mammalian circadian mutation tau. Science, 2000. 288(5465): p. 483–92. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 84.Gallego M and Virshup DM, Post-translational modifications regulate the ticking of the circadian clock. Nat Rev Mol Cell Biol, 2007. 8(2): p. 139–48. [DOI] [PubMed] [Google Scholar]
- 85.Meng QJ, et al. , Setting clock speed in mammals: the CK1 epsilon tau mutation in mice accelerates circadian pacemakers by selectively destabilizing PERIOD proteins. Neuron, 2008. 58(1): p. 78–88. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 86.Lee HM, et al. , The period of the circadian oscillator is primarily determined by the balance between casein kinase 1 and protein phosphatase 1. Proc Natl Acad Sci U S A, 2011. 108(39): p. 16451–6. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 87.Camacho F, et al. , Human casein kinase Idelta phosphorylation of human circadian clock proteins period 1 and 2. FEBS Lett, 2001. 489(2–3): p. 159–65. [DOI] [PubMed] [Google Scholar]
- 88.Eide EJ, et al. , Control of mammalian circadian rhythm by CKIepsilon-regulated proteasome-mediated PER2 degradation. Mol Cell Biol, 2005. 25(7): p. 2795–807. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 89.Shirogane T, et al. , SCFbeta-TRCP controls clock-dependent transcription via casein kinase 1-dependent degradation of the mammalian period-1 (Per1) protein. J Biol Chem, 2005. 280(29): p. 26863–72. [DOI] [PubMed] [Google Scholar]
- 90.Vanselow K, et al. , Differential effects of PER2 phosphorylation: molecular basis for the human familial advanced sleep phase syndrome (FASPS). Genes Dev, 2006. 20(19): p. 2660–72. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 91.Zhou M, et al. , A Period2 Phosphoswitch Regulates and Temperature Compensates Circadian Period. Mol Cell, 2015. 60(1): p. 77–88. [DOI] [PubMed] [Google Scholar]
- 92.Ohsaki K, et al. , The role of {beta}-TrCP1 and {beta}-TrCP2 in circadian rhythm generation by mediating degradation of clock protein PER2. J Biochem, 2008. 144(5): p. 609–18. [DOI] [PubMed] [Google Scholar]
- 93.Reischl S, et al. , Beta-TrCP1-mediated degradation of PERIOD2 is essential for circadian dynamics. J Biol Rhythms, 2007. 22(5): p. 375–86. [DOI] [PubMed] [Google Scholar]
- 94.Narasimamurthy R, et al. , CK1delta/epsilon protein kinase primes the PER2 circadian phosphoswitch. Proc Natl Acad Sci U S A, 2018. 115(23): p. 5986–5991. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 95.Meng QJ, et al. , Entrainment of disrupted circadian behavior through inhibition of casein kinase 1 (CK1) enzymes. Proc Natl Acad Sci U S A, 2010. 107(34): p. 15240–5. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 96.Masuda S, et al. , Mutation of a PER2 phosphodegron perturbs the circadian phosphoswitch. Proc Natl Acad Sci U S A, 2020. 117(20): p. 10888–10896. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 97.Tsuchiya Y, et al. , Involvement of the protein kinase CK2 in the regulation of mammalian circadian rhythms. Sci Signal, 2009. 2(73): p. ra26. [DOI] [PubMed] [Google Scholar]
- 98.Maier B, et al. , A large-scale functional RNAi screen reveals a role for CK2 in the mammalian circadian clock. Genes Dev, 2009. 23(6): p. 708–18. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 99.Oshima T, et al. , Cell-based screen identifies a new potent and highly selective CK2 inhibitor for modulation of circadian rhythms and cancer cell growth. Sci Adv, 2019. 5(1): p. eaau9060. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 100.Hirota T, et al. , High-throughput chemical screen identifies a novel potent modulator of cellular circadian rhythms and reveals CKIalpha as a clock regulatory kinase. PLoS Biol, 2010. 8(12): p. e1000559. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 101.Siepka SM, et al. , Circadian mutant Overtime reveals F-box protein FBXL3 regulation of cryptochrome and period gene expression. Cell, 2007. 129(5): p. 1011–23. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 102.Godinho SI, et al. , The after-hours mutant reveals a role for Fbxl3 in determining mammalian circadian period. Science, 2007. 316(5826): p. 897–900. [DOI] [PubMed] [Google Scholar]
- 103.Busino L, et al. , SCFFbxl3 controls the oscillation of the circadian clock by directing the degradation of cryptochrome proteins. Science, 2007. 316(5826): p. 900–4. [DOI] [PubMed] [Google Scholar]
- 104.Correia SP, et al. , The circadian E3 ligase complex SCF. Sci Rep, 2019. 9(1): p. 198. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 105.Hirano A, et al. , FBXL21 regulates oscillation of the circadian clock through ubiquitination and stabilization of cryptochromes. Cell, 2013. 152(5): p. 1106–18. [DOI] [PubMed] [Google Scholar]
- 106.Yoo SH, et al. , Competing E3 ubiquitin ligases govern circadian periodicity by degradation of CRY in nucleus and cytoplasm. Cell, 2013. 152(5): p. 1091–105. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 107.Lamia KA, et al. , AMPK regulates the circadian clock by cryptochrome phosphorylation and degradation. Science, 2009. 326(5951): p. 437–40. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 108.Harada Y, et al. , Ser-557-phosphorylated mCRY2 is degraded upon synergistic phosphorylation by glycogen synthase kinase-3 beta. J Biol Chem, 2005. 280(36): p. 31714–21. [DOI] [PubMed] [Google Scholar]
- 109.Kurabayashi N, et al. , DYRK1A and glycogen synthase kinase 3beta, a dual-kinase mechanism directing proteasomal degradation of CRY2 for circadian timekeeping. Mol Cell Biol, 2010. 30(7): p. 1757–68. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 110.Sahar S, et al. , Regulation of BMAL1 protein stability and circadian function by GSK3beta-mediated phosphorylation. PLoS One, 2010. 5(1): p. e8561. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 111.Spengler ML, et al. , A serine cluster mediates BMAL1-dependent CLOCK phosphorylation and degradation. Cell Cycle, 2009. 8(24): p. 4138–46. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 112.Yin L, et al. , Nuclear receptor Rev-erbalpha is a critical lithium-sensitive component of the circadian clock. Science, 2006. 311(5763): p. 1002–5. [DOI] [PubMed] [Google Scholar]
- 113.Iitaka C, et al. , A role for glycogen synthase kinase-3beta in the mammalian circadian clock. J Biol Chem, 2005. 280(33): p. 29397–402. [DOI] [PubMed] [Google Scholar]
- 114.Vielhaber E, et al. , Nuclear entry of the circadian regulator mPER1 is controlled by mammalian casein kinase I epsilon. Mol Cell Biol, 2000. 20(13): p. 4888–99. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 115.Akashi M, et al. , Control of intracellular dynamics of mammalian period proteins by casein kinase I epsilon (CKIepsilon) and CKIdelta in cultured cells. Mol Cell Biol, 2002. 22(6): p. 1693–703. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 116.Gallego M, Kang H, and Virshup DM, Protein phosphatase 1 regulates the stability of the circadian protein PER2. Biochem J, 2006. 399(1): p. 169–75. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 117.Partch CL, et al. , Posttranslational regulation of the mammalian circadian clock by cryptochrome and protein phosphatase 5. Proc Natl Acad Sci U S A, 2006. 103(27): p. 10467–10472. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 118.Cardone L, et al. , Circadian clock control by SUMOylation of BMAL1. Science, 2005. 309(5739): p. 1390–4. [DOI] [PubMed] [Google Scholar]
- 119.Petkau N, et al. , Acetylation of BMAL1 by TIP60 controls BRD4-P-TEFb recruitment to circadian promoters. Elife, 2019. 8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 120.Kaasik K, et al. , Glucose sensor O-GlcNAcylation coordinates with phosphorylation to regulate circadian clock. Cell Metab, 2013. 17(2): p. 291–302. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 121.Ripperger JA and Schibler U, Rhythmic CLOCK-BMAL1 binding to multiple E-box motifs drives circadian Dbp transcription and chromatin transitions. Nat Genet, 2006. 38(3): p. 369–74. [DOI] [PubMed] [Google Scholar]
- 122.Ueda HR, et al. , A transcription factor response element for gene expression during circadian night. Nature, 2002. 418(6897): p. 534–9. [DOI] [PubMed] [Google Scholar]
- 123.Menet JS, et al. , Nascent-Seq reveals novel features of mouse circadian transcriptional regulation. Elife, 2012. 1: p. e00011. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 124.Vollmers C, et al. , Circadian oscillations of protein-coding and regulatory RNAs in a highly dynamic mammalian liver epigenome. Cell Metab, 2012. 16(6): p. 833–45. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 125.Yoshitane H, et al. , CLOCK-controlled polyphonic regulation of circadian rhythms through canonical and noncanonical E-boxes. Mol Cell Biol, 2014. 34(10): p. 1776–87. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 126.Storch KF, et al. , Intrinsic circadian clock of the mammalian retina: importance for retinal processing of visual information. Cell, 2007. 130(4): p. 730–741. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 127.Kornmann B, et al. , Analysis of circadian liver gene expression by ADDER, a highly sensitive method for the display of differentially expressed mRNAs. Nucleic Acids Res, 2001. 29(11): p. E51–1. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 128.Panda S, et al. , Coordinated transcription of key pathways in the mouse by the circadian clock. Cell, 2002. 109(3): p. 307–20. [DOI] [PubMed] [Google Scholar]
- 129.Storch KF, et al. , Extensive and divergent circadian gene expression in liver and heart. Nature, 2002. 417(6884): p. 78–83. [DOI] [PubMed] [Google Scholar]
- 130.Zhang R, et al. , A circadian gene expression atlas in mammals: implications for biology and medicine. Proc Natl Acad Sci U S A, 2014. 111(45): p. 16219–24. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 131.Bass J, Circadian topology of metabolism. Nature, 2012. 491(7424): p. 348–56. [DOI] [PubMed] [Google Scholar]
- 132.Scheiermann C, Kunisaki Y, and Frenette PS, Circadian control of the immune system. Nat Rev Immunol, 2013. 13(3): p. 190–8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 133.Curtis AM, et al. , Circadian clock proteins and immunity. Immunity, 2014. 40(2): p. 178–86. [DOI] [PubMed] [Google Scholar]
- 134.Le Martelot G, et al. , Genome-wide RNA polymerase II profiles and RNA accumulation reveal kinetics of transcription and associated epigenetic changes during diurnal cycles. PLoS Biol, 2012. 10(11): p. e1001442. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 135.Xu Y, et al. , Long-Range Chromosome Interactions Mediated by Cohesin Shape Circadian Gene Expression. PLoS Genet, 2016. 12(5): p. e1005992. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 136.Kim YH, et al. , Rev-erbalpha dynamically modulates chromatin looping to control circadian gene transcription. Science, 2018. 359(6381): p. 1274–1277. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 137.Mermet J, et al. , Clock-dependent chromatin topology modulates circadian transcription and behavior. Genes Dev, 2018. 32(5–6): p. 347–358. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 138.Yeung J and Naef F, Rhythms of the Genome: Circadian Dynamics from Chromatin Topology, Tissue-Specific Gene Expression, to Behavior. Trends Genet, 2018. 34(12): p. 915–926. [DOI] [PubMed] [Google Scholar]
- 139.Pacheco-Bernal I, Becerril-Perez F, and Aguilar-Arnal L, Circadian rhythms in the three-dimensional genome: implications of chromatin interactions for cyclic transcription. Clin Epigenetics, 2019. 11(1): p. 79. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 140.Sawant OB, et al. , The circadian clock gene Bmal1 is required to control the timing of retinal neurogenesis and lamination of Muller glia in the mouse retina. FASEB J, 2019. 33(8): p. 8745–8758. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 141.Mure LS, et al. , Diurnal transcriptome atlas of a primate across major neural and peripheral tissues. Science, 2018. 359(6381). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 142.Stephan FK and Zucker I, Circadian rhythms in drinking behavior and locomotor activity of rats are eliminated by hypothalamic lesions. Proc Natl Acad Sci U S A, 1972. 69(6): p. 1583–6. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 143.Moore RY and Eichler VB, Loss of a circadian adrenal corticosterone rhythm following suprachiasmatic lesions in the rat. Brain Res, 1972. 42(1): p. 201–6. [DOI] [PubMed] [Google Scholar]
- 144.Ralph MR, et al. , Transplanted suprachiasmatic nucleus determines circadian period. Science, 1990. 247(4945): p. 975–8. [DOI] [PubMed] [Google Scholar]
- 145.Eastman CI, Mistlberger RE, and Rechtschaffen A, Suprachiasmatic nuclei lesions eliminate circadian temperature and sleep rhythms in the rat. Physiol Behav, 1984. 32(3): p. 357–68. [DOI] [PubMed] [Google Scholar]
- 146.Kawamura H and Ibuka N, The search for circadian rhythm pacemakers in the light of lesion experiments. Chronobiologia, 1978. 5(1): p. 69–88. [PubMed] [Google Scholar]
- 147.Cassone VM, et al. , Comparative anatomy of the mammalian hypothalamic suprachiasmatic nucleus. J Biol Rhythms, 1988. 3(1): p. 71–91. [DOI] [PubMed] [Google Scholar]
- 148.Bunning E, Das weiterlaufen der “physiologischen uhr” in sugerdarm ohne zentrale steuerung. Naturwissenschaften, 1958. 45: p. 68. [Google Scholar]
- 149.Andrews RV and Folk GE Jr., Circadian Metabolic Patterns in Cultured Hamster Adrenal Glands. Comp Biochem Physiol, 1964. 11: p. 393–409. [DOI] [PubMed] [Google Scholar]
- 150.Andrews RV and Shiotsuka R, The effect of actinomycin D on the in vitro adrenal secretory rhythm of the hamster. Comp Biochem Physiol, 1970. 36(2): p. 353–63. [DOI] [PubMed] [Google Scholar]
- 151.Andrews RV, Circadian rhythms in adrenal organ cultures. Gegenbaurs Morphol Jahrb, 1971. 117(1): p. 89–98. [PubMed] [Google Scholar]
- 152.Shiotsuka R, Jovonovich J, and Jovonovich J, Circadian and ultradian corticosterone rhythms in adrenal organ cultures. Chronobiologia, 1974. 1 Suppl 1: p. 109–21. [PubMed] [Google Scholar]
- 153.Tharp GD and Folk GE Jr., Rhythmic Changes in Rate of the Mammalian Heart and Heart Cells during Prolonged Isolation. Comp Biochem Physiol, 1965. 14: p. 255–73. [DOI] [PubMed] [Google Scholar]
- 154.Langner R and Rensing L, Circadian rhythm of oxygen consumption in rat liver suspension culture: changes of pattern. Z Naturforsch B, 1972. 27(9): p. 1117–8. [DOI] [PubMed] [Google Scholar]
- 155.Tei H, et al. , Circadian oscillation of a mammalian homologue of the Drosophila period gene. Nature, 1997. 389(6650): p. 512–6. [DOI] [PubMed] [Google Scholar]
- 156.Sun ZS, et al. , RIGUI, a putative mammalian ortholog of the Drosophila period gene. [DOI] [PubMed] [Google Scholar]
- 157.Stratmann M and Schibler U, Properties, entrainment, and physiological functions of mammalian peripheral oscillators. J Biol Rhythms, 2006. 21(6): p. 494–506. [DOI] [PubMed] [Google Scholar]
- 158.Nagoshi E, et al. , Circadian gene expression in individual fibroblasts: cell-autonomous and self-sustained oscillators pass time to daughter cells. Cell, 2004. 119(5): p. 693–705. [DOI] [PubMed] [Google Scholar]
- 159.Welsh DK, et al. , Bioluminescence imaging of individual fibroblasts reveals persistent, independently phased circadian rhythms of clock gene expression. Curr Biol, 2004. 14(24): p. 2289–95. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 160.Mohawk JA, Green CB, and Takahashi JS, Central and peripheral circadian clocks in mammals. Annu Rev Neurosci, 2012. 35: p. 445–62. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 161.Hastings MH, Reddy AB, and Maywood ES, A clockwork web: circadian timing in brain and periphery, in health and disease. Nat Rev Neurosci, 2003. 4(8): p. 649–61. [DOI] [PubMed] [Google Scholar]
- 162.Gerber A, et al. , Blood-borne circadian signal stimulates daily oscillations in actin dynamics and SRF activity. Cell, 2013. 152(3): p. 492–503. [DOI] [PubMed] [Google Scholar]
- 163.Morf J and Schibler U, Body temperature cycles: gatekeepers of circadian clocks. Cell Cycle, 2013. 12(4): p. 539–40. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 164.Freedman MS, et al. , Regulation of mammalian circadian behavior by non-rod, non-cone, ocular photoreceptors. Science, 1999. 284(5413): p. 502–4. [DOI] [PubMed] [Google Scholar]
- 165.Panda S, et al. , Melanopsin (Opn4) requirement for normal light-induced circadian phase shifting. Science, 2002. 298(5601): p. 2213–6. [DOI] [PubMed] [Google Scholar]
- 166.Guler AD, et al. , Melanopsin cells are the principal conduits for rod-cone input to non-image-forming vision. Nature, 2008. 453(7191): p. 102–5. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 167.Ebling FJ, The role of glutamate in the photic regulation of the suprachiasmatic nucleus. Prog Neurobiol, 1996. 50(2–3): p. 109–32. [DOI] [PubMed] [Google Scholar]
- 168.Jones JR, Tackenberg MC, and McMahon DG, Manipulating circadian clock neuron firing rate resets molecular circadian rhythms and behavior. Nat Neurosci, 2015. 18(3): p. 373–5. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 169.Shearman LP, et al. , Two period homologs: circadian expression and photic regulation in the suprachiasmatic nuclei. Neuron, 1997. 19(6): p. 1261–9. [DOI] [PubMed] [Google Scholar]
- 170.Shigeyoshi Y, et al. , Light-induced resetting of a mammalian circadian clock is associated with rapid induction of the mPer1 transcript. Cell, 1997. 91(7): p. 1043–53. [DOI] [PubMed] [Google Scholar]
- 171.Albrecht U, et al. , A differential response of two putative mammalian circadian regulators, mper1 and mper2, to light. Cell, 1997. 91(7): p. 1055–64. [DOI] [PubMed] [Google Scholar]
- 172.Brown SA, et al. , Rhythms of mammalian body temperature can sustain peripheral circadian clocks. Curr Biol, 2002. 12(18): p. 1574–83. [DOI] [PubMed] [Google Scholar]
- 173.Buhr ED, Yoo SH, and Takahashi JS, Temperature as a universal resetting cue for mammalian circadian oscillators. Science, 2010. 330(6002): p. 379–85. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 174.Ruby NF, et al. , Suprachiasmatic nuclei influence torpor and circadian temperature rhythms in hamsters. Am J Physiol, 1989. 257(1 Pt 2): p. R210–5. [DOI] [PubMed] [Google Scholar]
- 175.Kornmann B, et al. , System-driven and oscillator-dependent circadian transcription in mice with a conditionally active liver clock. PLoS Biol, 2007. 5(2): p. e34. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 176.Reinke H, et al. , Differential display of DNA-binding proteins reveals heat-shock factor 1 as a circadian transcription factor. Genes Dev, 2008. 22(3): p. 331–45. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 177.Tamaru T, et al. , Synchronization of circadian Per2 rhythms and HSF1-BMAL1:CLOCK interaction in mouse fibroblasts after short-term heat shock pulse. PLoS One, 2011. 6(9): p. e24521. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 178.Morf J, et al. , Cold-inducible RNA-binding protein modulates circadian gene expression posttranscriptionally. Science, 2012. 338(6105): p. 379–83. [DOI] [PubMed] [Google Scholar]
- 179.Gotic I, et al. , Temperature regulates splicing efficiency of the cold-inducible RNA-binding protein gene Cirbp. Genes Dev, 2016. 30(17): p. 2005–17. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 180.Stokkan KA, et al. , Entrainment of the circadian clock in the liver by feeding. Science, 2001. 291(5503): p. 490–3. [DOI] [PubMed] [Google Scholar]
- 181.Damiola F, et al. , Restricted feeding uncouples circadian oscillators in peripheral tissues from the central pacemaker in the suprachiasmatic nucleus. Genes Dev, 2000. 14(23): p. 2950–61. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 182.Welz PS, et al. , BMAL1-Driven Tissue Clocks Respond Independently to Light to Maintain Homeostasis. Cell, 2019. 177(6): p. 1436–1447.e12. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 183.Buhr ED, et al. , Neuropsin (OPN5) Mediates Local Light-Dependent Induction of Circadian Clock Genes and Circadian Photoentrainment in Exposed Murine Skin. Curr Biol, 2019. 29(20): p. 3478–3487 e4. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 184.Kofuji P, et al. , Intrinsically Photosensitive Retinal Ganglion Cells (ipRGCs) Are Necessary for Light Entrainment of Peripheral Clocks. PLoS One, 2016. 11(12): p. e0168651. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 185.Tanioka M, et al. , Molecular clocks in mouse skin. J Invest Dermatol, 2009. 129(5): p. 1225–31. [DOI] [PubMed] [Google Scholar]
- 186.Buhr ED and Van Gelder RN, Local photic entrainment of the retinal circadian oscillator in the absence of rods, cones, and melanopsin. Proc Natl Acad Sci U S A, 2014. 111(23): p. 8625–30. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 187.Buhr ED, et al. , Neuropsin (OPN5)-mediated photoentrainment of local circadian oscillators in mammalian retina and cornea. Proc Natl Acad Sci U S A, 2015. 112(42): p. 13093–8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 188.Pittendrigh CS, Temporal organization: reflections of a Darwinian clock-watcher. Annu Rev Physiol, 1993. 55: p. 16–54. [DOI] [PubMed] [Google Scholar]
- 189.Reddy P, et al. , Molecular analysis of the period locus in Drosophila melanogaster and identification of a transcript involved in biological rhythms. Cell, 1984. 38(3): p. 701–10. [DOI] [PubMed] [Google Scholar]
- 190.Konopka RJ and Benzer S, Clock mutants of Drosophila melanogaster. Proc Natl Acad Sci U S A, 1971. 68(9): p. 2112–6. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 191.Bargiello TA, Jackson FR, and Young MW, Restoration of circadian behavioural rhythms by gene transfer in Drosophila. Nature, 1984. 312(5996): p. 752–4. [DOI] [PubMed] [Google Scholar]
- 192.Adam D, Core Concept: Emerging science of chronotherapy offers big opportunities to optimize drug delivery. Proc Natl Acad Sci U S A, 2019. 116(44): p. 21957–21959. [DOI] [PMC free article] [PubMed] [Google Scholar]


