Table 4.
List of known interaction sites deposited in PBD protein structures, located in close proximity to post-translationally modified amino acid residues found in patients with breast and ovaria cancer.
| Protein entry name | PDB ID | Ligand | Locus of interaction | References | Structure illustration |
|---|---|---|---|---|---|
| A1AT | 6HX4 | Fab-fragment | R199–Q215 and S362–K368 | 76 | 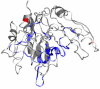 |
| 7NPL | Glycerol | V374 and L251 | 77 | ||
| 7NPK | Glycerol | S118 and L51 | 77 | ||
| 1EZX | Trypsin | S290–M335 | 78 | ||
| 1OPH | Trypsinogen | G349–P361 | 79 | ||
| 3CWM | Citrate | L241 and L288 | 80 | ||
| 7AEL | Sulfate anion | L298 | 81 | ||
| ALBU | 3B9M | Myristate | A158 and R186 | 82 | 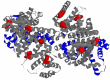 |
| 4Z69 | Palmitic acid | L345, C437 and A449 | 83 | ||
| 2ESG | IgA1 | P35–R144 | 84 | ||
| 2VUE | 4Z,15E-bilirubin-IX-alpha | R114, P127 and L182–K190 | 85 | ||
| APOA1 | 3R2P | Homodimer | R84–E111 and H162–A181 | 86 | 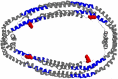 |
| 3K2S | Phospholipids | Whole seguence | 87 | ||
| TRFE | 3S9N | Transferrin receptor protein | L86–N90, R338–G343 and S362–N375 | 88 | 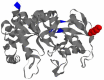 |
| 3S9M | Iron-cation | L242 | 88 |
Blue color indicates sites of interaction with ligands, red color designates amino acids with PTM found in our study.
