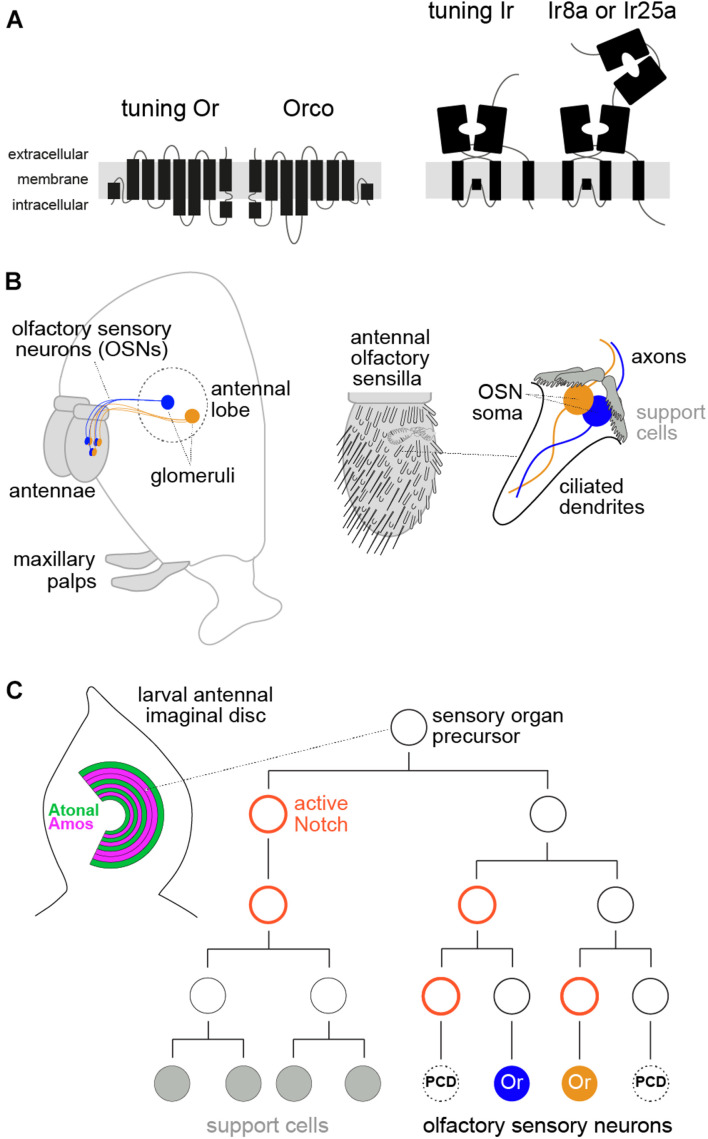
FIGURE 1.
Molecular, anatomical, and developmental properties of the peripheral olfactory system in D. melanogaster. (A) Schematic of the two main insect olfactory receptor families. Odorant receptors (Ors) are seven transmembrane domain proteins that form heteromeric odor-gated ion channels composed of subunits of a ligand-specific (“tuning”) receptor and a co-receptor, Orco. Ionotropic receptors (Irs) are distantly related to ionotropic glutamate receptors, and function as odor-gated channel complexes composed of tuning Ir subunits and co-receptors (Ir8a or Ir25a). (B) Left: schematic of the D. melanogaster head (facing left) illustrating the main olfactory organs (antennae and maxillary palps, gray shading) and connectivity of two populations of olfactory sensory neurons (OSNs) to the antennal lobe in the brain. Right: schematic of the antenna, which is covered with diverse classes of sensory sensilla; the cellular organization of one sensillum, housing two OSNs, is shown on the far right (see text). (C) Left: schematic of the larval antennal imaginal disc, showing the concentric arcs of cells where different sensory organ precursors (SOP) are born. Amos- and Atonal-positive arcs give rise to OSN lineages expressing Ors and Irs, respectively, while other patterning determinants (not shown) are thought to specify SOP identity for different sensilla subtypes. Right: a simplified developmental lineage of an SOP producing a sensillum class with two OSNs. Two other potential neurons are removed by programmed cell death (PCD). Delta/Notch signaling determines the asymmetry of cell divisions, while many other patterning factors (not shown) are involved in specifying cell identity, encompassing both receptor expression and glomerular targeting of different OSNs (see text).