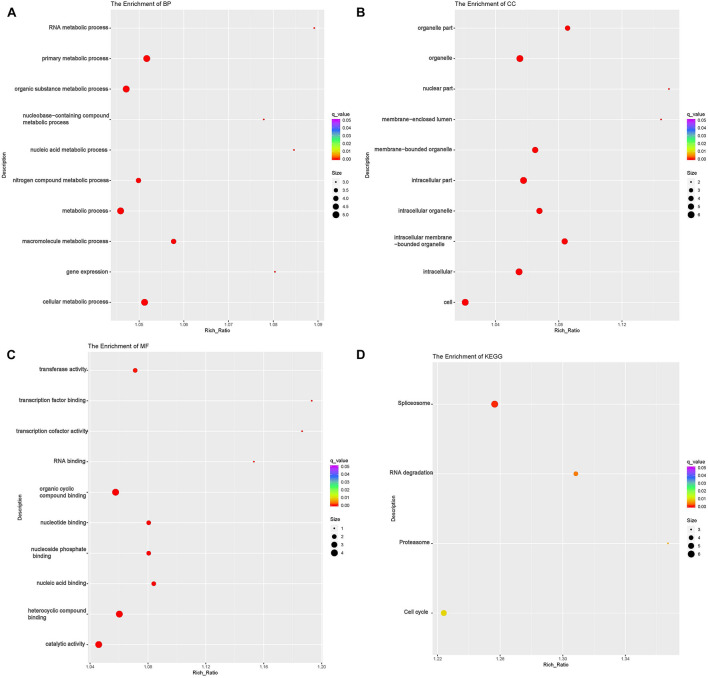
FIGURE 5.
Functional annotation of predicted targets of differentially expressed (DE) up-regulated long non-coding RNAs (lncRNA) with the top 10 enrichment scores of biological processes (BP) (A), cellular component (CC) (B), molecular function (MF) (C), and KEGG pathways (D). The abscissa is the rich ration [(DE genes of the term or pathway/all the DE genes)/(all the annotated gene of the term or pathway/all the annotated gene)]; the Y-axis shows the terms enriched. A higher rich ration correlates with lower p-values. Circle size represents the number of enriched genes, and the color indicates the degree of enrichment, with red representing the highest degree of enrichment.