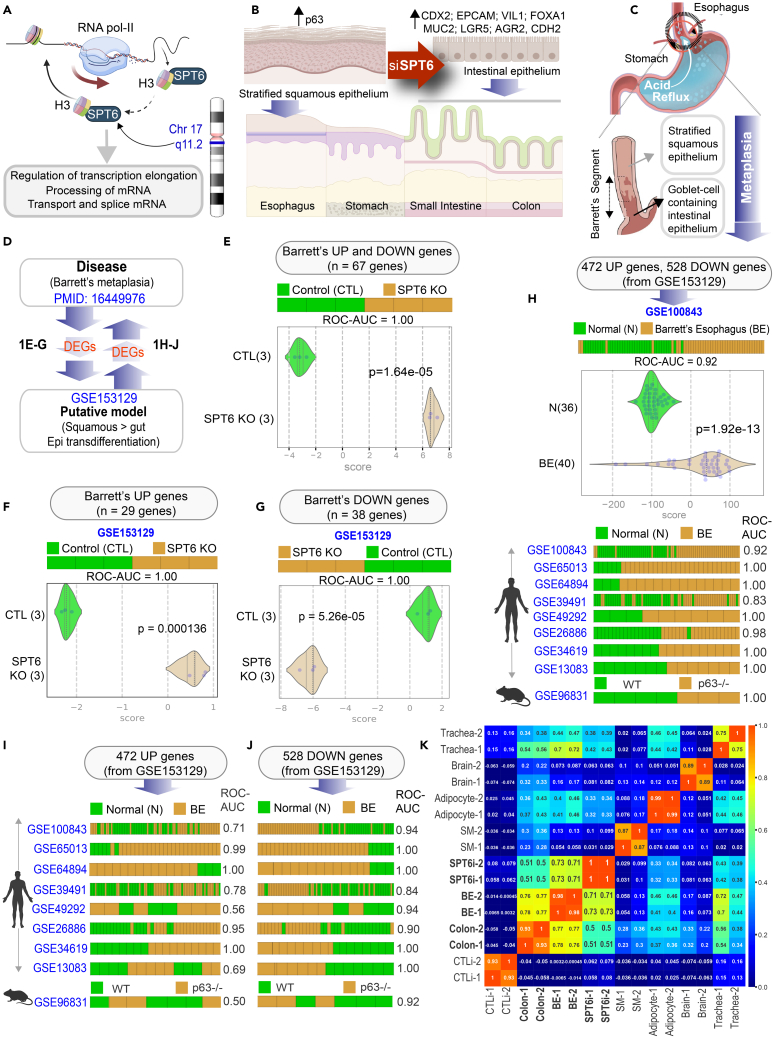
Figure 1.
Keratinocyte stem cells depleted of SPT6 trans-differentiates into gut lineage that resembles Barrett’s metaplasia
(A) Schematic summarizes the chromosomal location of SPT6 and its known functions in transcriptional elongation and mRNA processing. SPT6 coordinates nucleosome dis- and re-assembly, transcriptional elongation, and mRNA processing. SPT6 is a conserved factor that controls transcription and chromatin structure across the genome.
(B) Schematic summarizing the key findings in gene expression and epithelial morphology observed and reported earlier (Li et al., 2021) upon depletion of SPT6 in keratinocyte stem cells by siRNA (Li et al., 2021). While control keratinocytes formed stratified squamous epithelium, siRNA-mediated transient depletion of SPT6 in keratinocytes (SPT6i) grew as “intestine-like” monolayers.
(C) Schematic showing the only known human pathophysiologic context in which stratified squamous epithelium is known to be replaced by “intestine-like” epithelium.
(D) Summary of computational approach used in (E-J).
(E–G) Differentially expressed genes (DEGs; see Table S1) in Barrett’s metaplasia vs. normal esophagus were used to rank order control (CTLi) and SPT6-depleted samples (SPT6i), either using UP genes alone (F), DOWN-genes alone (G) or both UP and DOWN signatures together (E). Results are presented as bar (top) and violin (bottom) plots. ROC-AUC in all cases reflects a perfect strength of classification (1.00). Welch’s two sample unpaired t test is performed on the composite gene signature score to compute the p values.
(H–J) Differentially expressed genes (DEGs; see Data S1) between control (CTLi) and SPT6-depleted (SPT6i) samples were used to rank order normal (N) from Barrett’s esophageal (BE) samples across 9 publicly available independent cohorts (8 human, 1 mouse), either using UP genes alone (I), DOWN-genes alone (J), or both UP and DOWN signatures together (H). See also Figure S1 for violin plots for each dataset. ROC-AUC in each case is annotated on the right side of the corresponding bar plots.
(K) Pearson correlation matrix showing clustering of control (CTLi) and SPT6-depleted (SPT6i) gene expression signatures with the brain, colon, BE, adipocyte, trachea, and skeletal muscle. Two distinct RNA-Seq samples of adult origin are shown for each tissue (see Table S2 for the list of datasets used to generate the matrix).