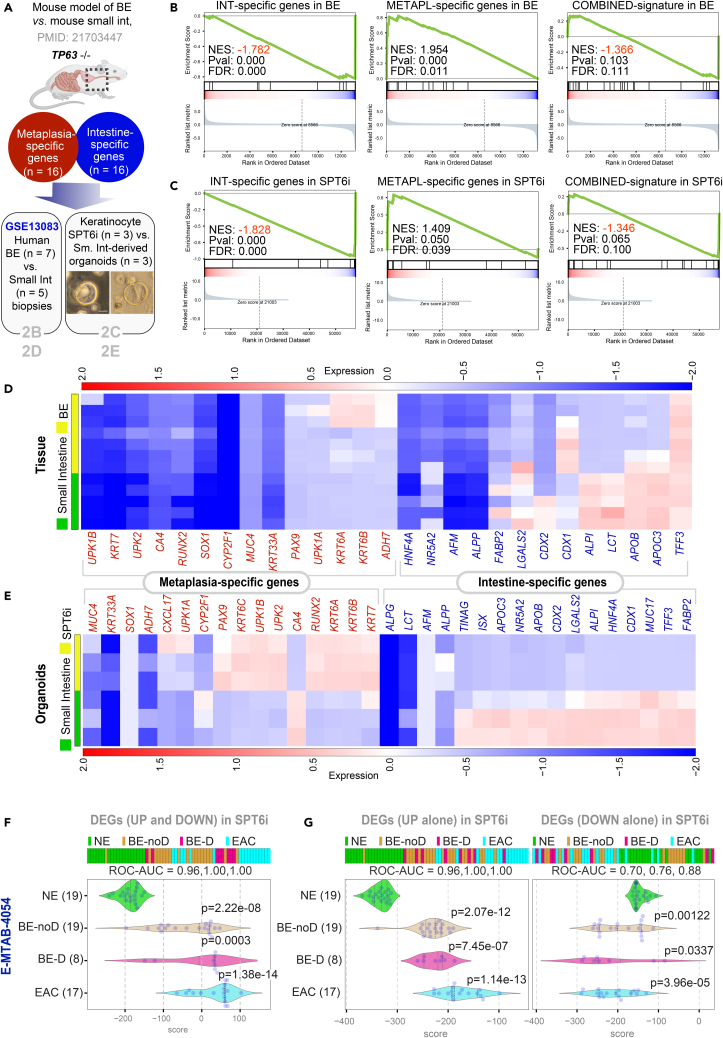
Figure 2.
Downregulation of SPT6 enriches metaplasia-specific genes
(A–E) Schematic in (A) summarizing the workflow for distinguishing BE from intestine. Using TP63−/− as a strategy to induce BE in mice, a prior study showed that compared to intestinal tissues, BE tissue was enriched in a 16-gene metaplasia-specific signature and de-enriched in a 16-gene intestine-specific signature (see Table S3 for the list of genes). These gene sets were analyzed for enrichment (Gene Set Enrichment Analysis – GSEA pre-ranked analysis) in human BE vs. small intestine tissues (GSE13083; B) and SPT6-depleted (SPT6i) vs. small intestine-derived organoids (C; scale bar = 100 μm). Heatmaps (D and E) display the levels of expression of the individual metaplasia-specific and intestine-specific genes in the BE vs. small intestine tissues (D) and organoids SPT6-depleted keratinocyte (SPT6i) vs. small intestine-derived organoids (E).
(F and G) Differentially expressed genes (DEGs; see Data S1) between control (CTLi) and SPT6-depleted (SPT6i) samples were used to rank order normal squamous esophagus (NE) from non-dysplastic Barrett’s esophagus (BE-noD), dysplastic BE (BE-D), and esophageal adenocarcinoma (EAC; n = 12) samples in an RNA seq dataset [E-MTAB-4054] (Maag et al., 2017), either using UP genes alone (G; left), DOWN-genes alone (G; right), or both UP and DOWN signatures together (F). Numbers in parenthesis indicate the number of samples. ROC-AUC in each case is annotated below the bar plots. Welch’s two sample unpaired t test is performed on the composite gene signature score to compute the p values. In multi-group setting, each group is compared to the NE control group and only significant p values are displayed.