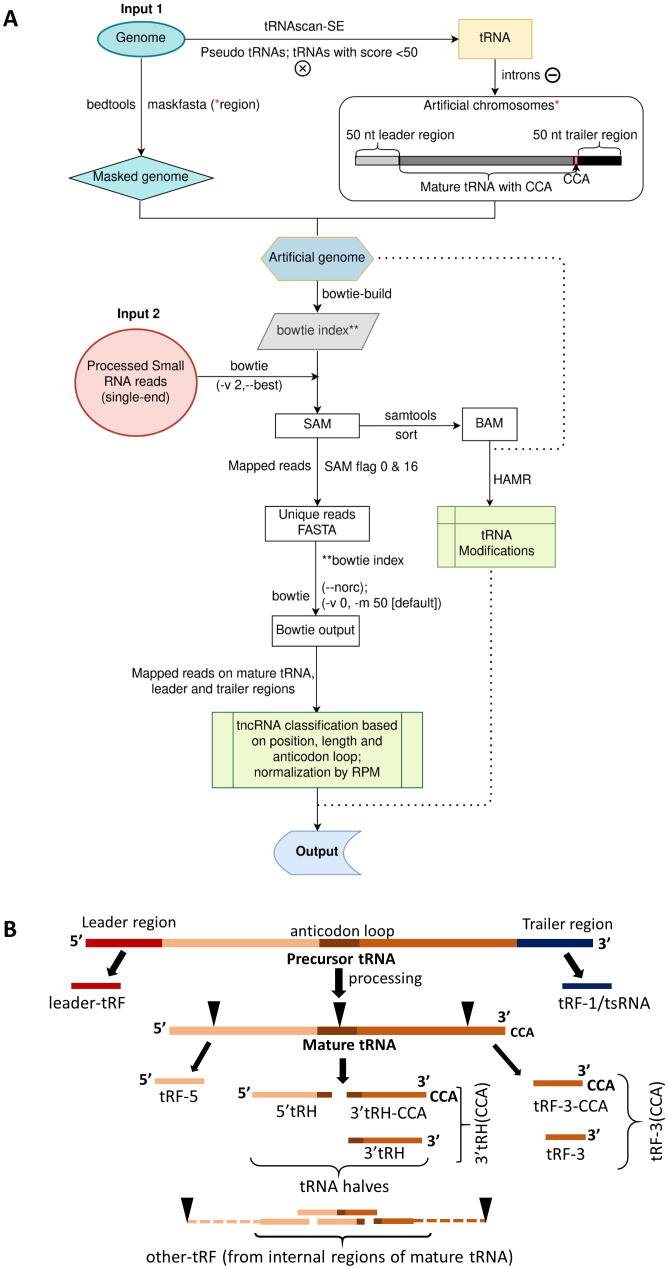
Fig. 1.
Workflow for the identification of transfer-RNA-derived non-coding RNAs (tncRNAs). A Genome and filtered reads are provided as inputs to the pipeline. For avoiding ambiguous reads from non-tRNA regions, an artificial genome is created by masking the genuine tRNA gene region with 50 nt upstream and downstream in the genome and adding them as artificial chromosomes* (*as represented in the box). Only those reads are classified as tncRNAs that are mapped exclusively to the artificial chromosomes. tncRNAs are classified based on tRNA origin, and site of cleavage. B Biogenesis of distinct tncRNA subtypes generated from the excision of pre-tRNA, and mature tRNA. pre-tRNA gives rise to leader tRF and tRF-1/tsRNA, whereas mature tRNA generates tRF-5, tRF-3 (with or without CCA), 5′tRH, 3′tRH (with or without CCA), and other tRF (derived from internal portions excluding extreme ends of mature tRNA denoted by dashed lines).