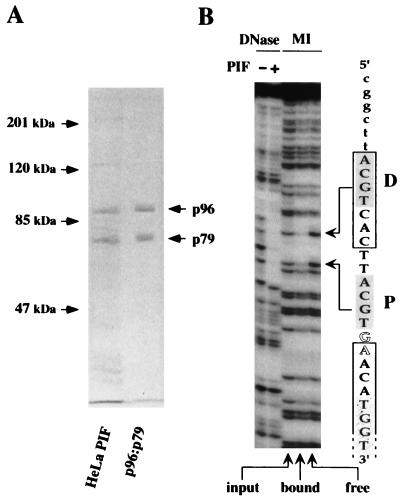
FIG. 4.
Characterization of recombinant PIF subunits expressed from separate baculovirus vectors. (A) SDS-PAGE analysis of purified recombinant PIF. DNA-affinity purified PIF from HeLaS3 cells was compared to purified recombinant PIF p96-p79, expressed in insect cells by using baculovirus vectors. The positions of the p96 and p79 subunits are indicated by arrows. (B) DNase I protection and methylation interference patterns of rPIF binding to a duplex DNA fragment containing the minimal replication origin, in which the top strand, as represented in Fig. 1, was 32P labeled at its 3′ end. The sequence through this part of the origin is indicated on the right. Lowercase letters indicate flanking vector DNA sequence. PIF half-sites are shaded, the consensus ATF site is boxed, bubble nucleotides are shown in outline, and the open-ended box indicates the start of the NS1 footprint, in which the first TGGT motif is stippled. Sequences protected by rPIF from DNase I digestion are shown in the left lanes. The right three lanes, labeled MI, show methylation interference profiles: input indicates piperidine cleavage products of the methylated DNA sample used in the binding assay; bound indicates cleaved DNA from the retarded rPIF-DNA complex; and free indicates free probe which was not shifted by the binding reaction. Methylated residues which impair rPIF binding are indicated by arrows.