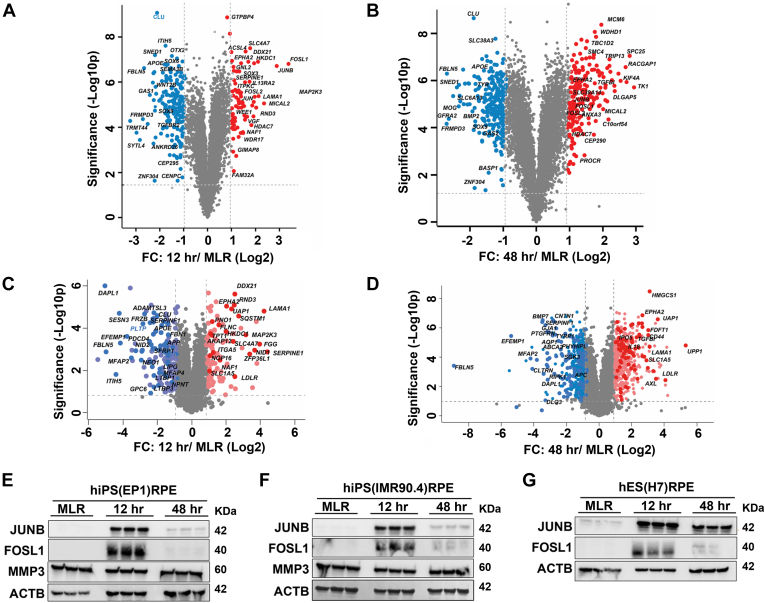
Fig. 3.
Dissociation-induced EMT alters malignancy-associated mesenchymal proteins and RPE signature proteins.A and B, volcano plot illustration of differentially expressed proteins during RPE-EMT that were identified by TMT labeling approach. C and D, volcano plot illustration of differentially expressed proteins during RPE-EMT were validated by direct-DIA approach. Pairwise comparison between (A) group I (12 h/MLR) and (B) group II (48 h/MLR). Expression fold changes (t test difference log 2, permutation-based FDR: 0.01, S0: 0.001) were calculated and plotted against permutation-based FDR of 0.01 cP-valued −log10 p value. Pairwise comparison between 12 h/MLR (C) and 48 h/MLR (D). E–G, Western blot validation of differentially regulated proteins identified from TMT labeling. Differentially expressed selective proteins identified from TMT labeling approach were validated in (E) hiPS-RPE (EP1), (F) hiPS-RPE (IMR90.4), and (G) hES (H7) that undergo EMT (12 and 48 h). DIA, data-independent acquisition; EMT, epithelial-to-mesenchymal transition; FDR, false discovery rate; hES, human-embryonic stem cell; hiPS, human-induce pluripotent stem cell; RPE, retinal pigment epithelium; TMT, tandem mass tag.