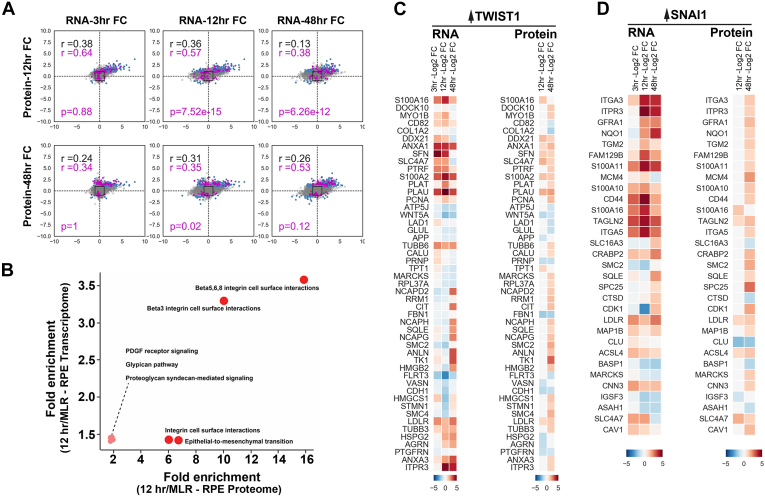
Fig. 4.
Integrated transcriptome and proteome changes show the correlation of malignancy-associated EMT and RPE-EMT.A, scatter plot showing the level of correlation between RNA (RNA-Seq; 3, 12, and 48 h) and protein (TMT-labeled proteomics; 12 and 48 h) expression changes during RPE-EMT. Gray dots represent shared genes/proteins between RNA-Seq and proteomics datasets. Black squares represent the threshold of log2FC, and blue dots were genes/proteins with log2FC larger than 1. Each dot in magenta represents a gene/protein that were altered from both hRPE EMT and malignancy-associated cells that were overexpressed with TWIST1 and SNAI1. B, scatter plot representation of pathway enrichment analysis that performed using functional enrichment tool (FUNrich). C, heat map representing the factors that were significantly altered from both dissociation-induced hRPE EMT model and TWIST1 overexpression-associated cancer EMT model at both RNA and protein levels. D, heat map showed the significantly altered factors from both dissociation-induced hRPE EMT model and SNAI1 overexpression-associated cancer EMT model at both RNA and protein levels. EMT, epithelial-to-mesenchymal transition; FC, fold change; RPE, retinal pigment epithelium; TMT, tandem mass tag.