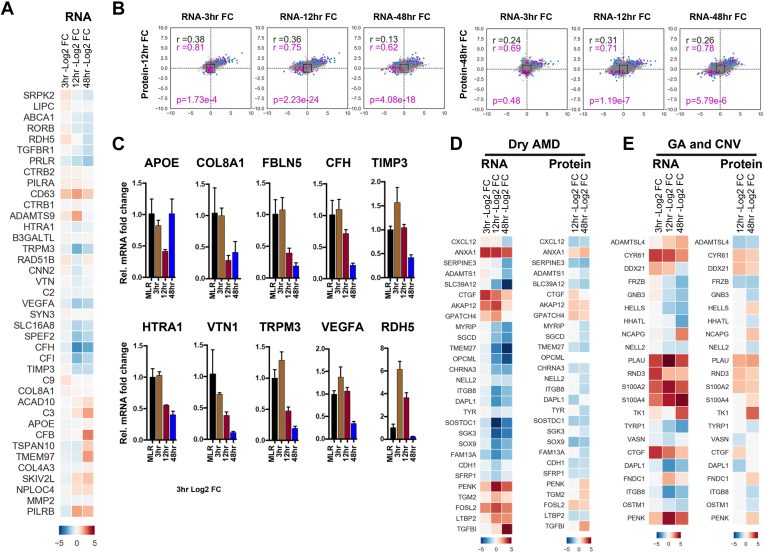
Fig. 5.
Integrative RPE-EMT transcriptome and proteome signature correlates with known AMD-associated risk factors.A, heat map representing the log2 fold change of AMD-associated risk variants that were significantly altered in RPE-EMT transcriptome study. B, scatter plot represents the correlation between changes in RNA (RNA-Seq; 3, 12, and 48 h) and protein (TMT-labeled proteomics; 12 and 48 h) levels following EMT induction. Dots in gray represent shared genes/proteins between RNA-Seq and proteomics datasets. Black squares represent the threshold of log2FC, and blue dots were genes/proteins with log2FC larger than 1. Dots in magenta represent the AMD-associated risk factors that were found in our RNA-Seq and proteomics study. C, differential expression of altered AMD-associated risk factors in RPE-EMT was measured by quantitative RT-PCR after enzymatic dissociation of monolayers into single cells. Data were normalized by the expression levels in monolayer control conditions. D and E, heat maps represent the list of common RPE-EMT genes and proteins that were enriched in a previously reported transcriptome study of AMD donor tissues. AMD, age-related macular degeneration; EMT, epithelial-to-mesenchymal transition; FC, fold change; RPE, retinal pigment epithelium; TMT, tandem mass tag.