Figure 3.
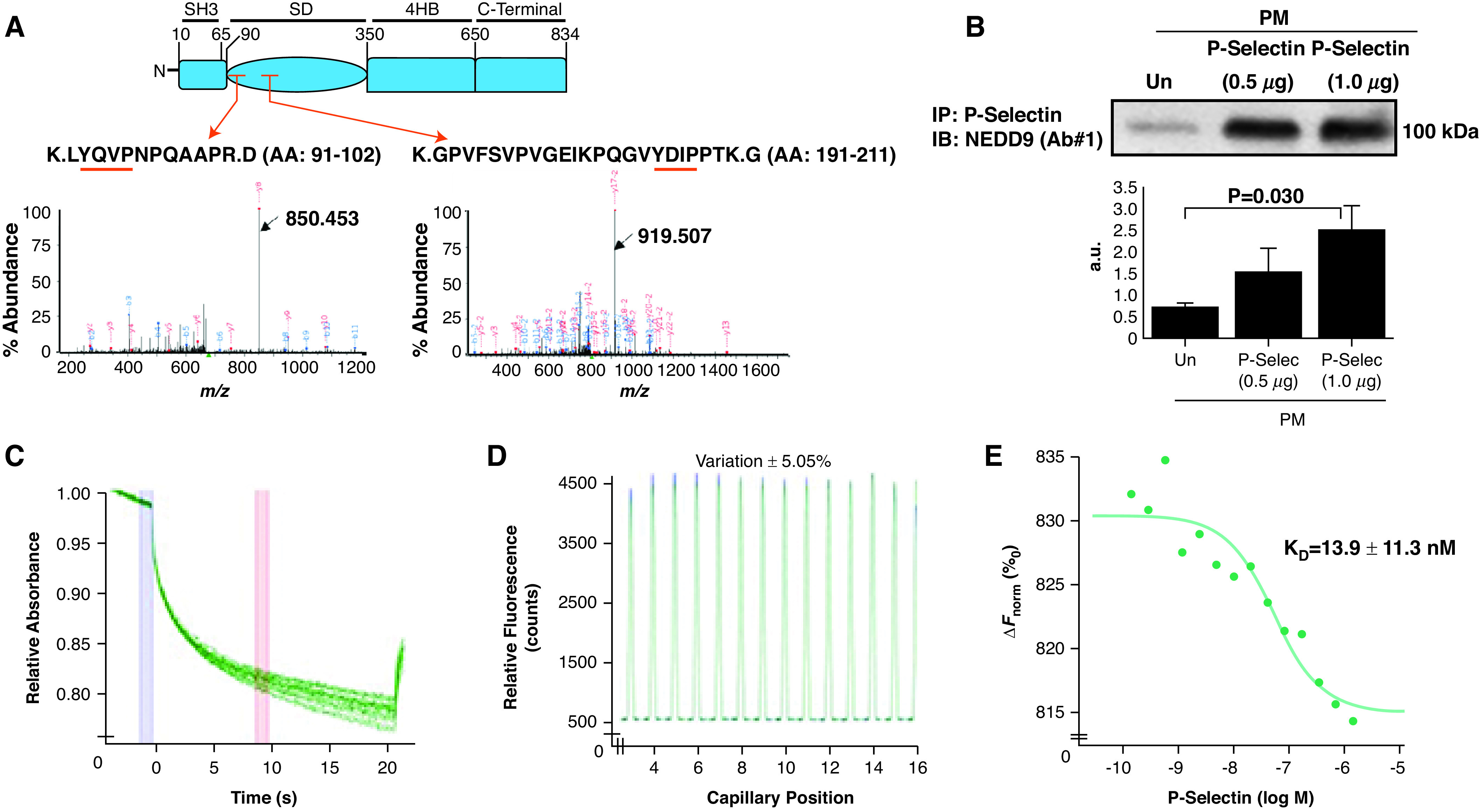
P-Selec (P-selectin) binds the NEDD9 substrate domain, and NEDD9 modulates platelet–endothelial adhesion. (A) Plasma-membrane fractions incubated with recombinant P-Selec for 1 hour were immunoprecipitated with an anti–P-Selec antibody (Ab). The fragmented ion MS spectra for each of the two detected NEDD9 peptide sequences, both within the substrate domain, are shown (black arrows): K.LYQVPNPQAAPR.D (amino acid [AA], 91–102) (m/z 677.36735 at retention time of 28.1 s) (NEDD9 peptide sequence 1) and K.GPVFSVPVGEIKPQGVYDIPPTK.G (AA, 191–211) (m/z 808.77731 at retention time 35.5 s) (NEDD9 peptide sequence 2) (n = 2 replicates for n = 2 iterations). The red arrows indicate the peptide location within the NEDD9 substrate domain (SD). The red underlining indicates a tyrosine-x-x-proline (YxxP) sequence, where x is another amino acid. (B) Human pulmonary-endothelial-cell plasma-membrane fractions were incubated with exogenous P-Selec, and coimmunoprecipitation was performed using an anti–P-Selec Ab followed by anti–NEDD9 immunoblot NEDD9. Varying concentrations of P-Selec (ligand) (2 μM–0.5 nM) were coincubated with fluorescently labeled NEDD9 (receptor) (20 nM), and microscale thermophoresis was performed to assess macromolecular interactions between these proteins. (C) Raw fluorescence tracings, (D) a capillary scan, and (E) a dose titration curve show a definitive protein–protein interaction between the receptor and ligand (Kd = 13.9 ± 11.3 nM) (n = 2). Data are presented as the mean ± SE. Representative immunoblot and titration curve are shown. a.u. = arbitrary units; ΔFnorm = change in normalized fluorescence; IB = immunoblot; IP = immunoprecipitation; M = molar; MS = mass spectrometry; m/z = mass-to-charge ratio; PM = plasma membrane; Un = untreated.
