Figure 4.
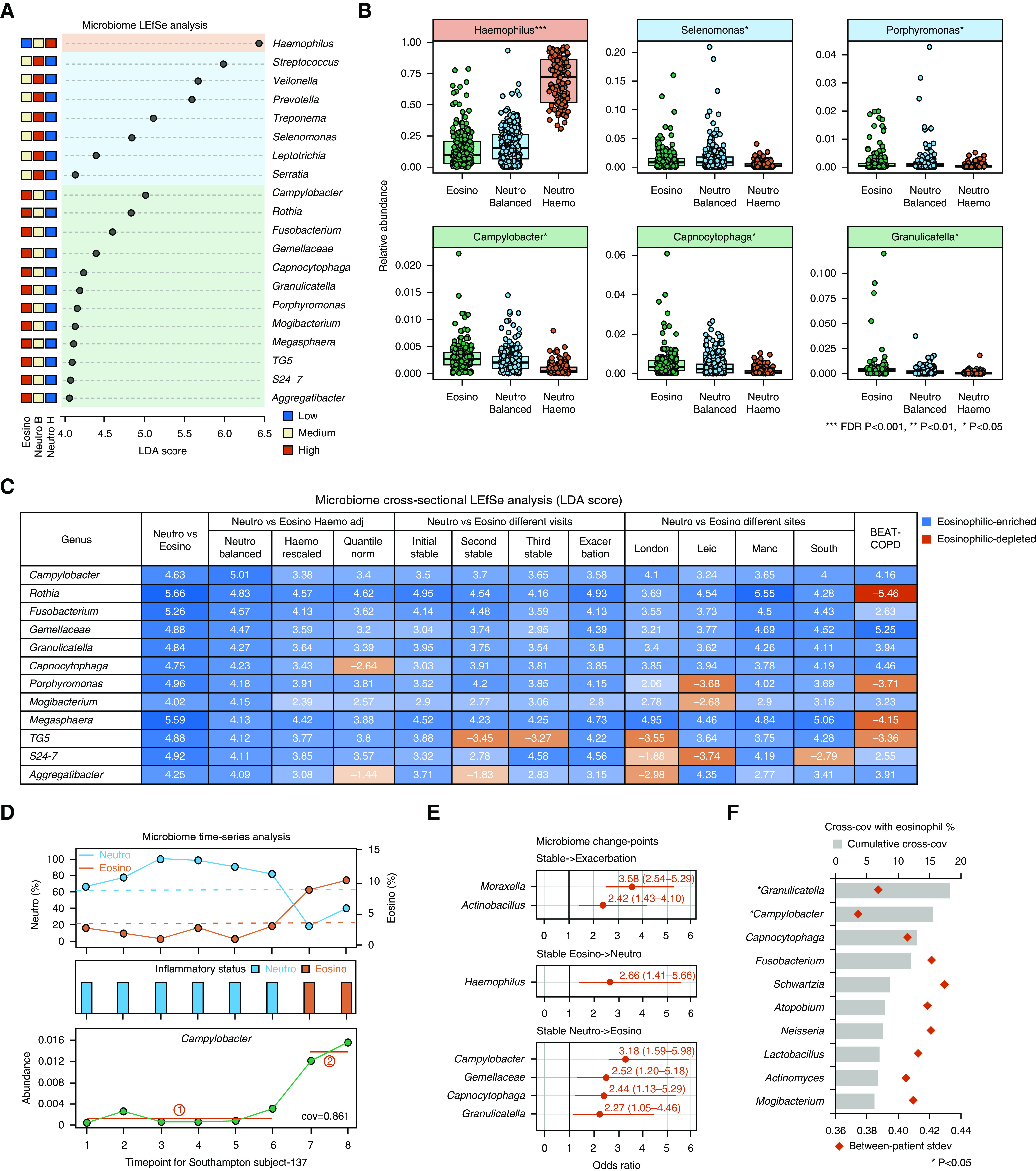
Specific nondominant microbiome genera were associated with eosinophilia. (A) Linear discriminant analysis (LDA) effect size (LEfSe) analysis showing microbiome genera specifically enriched in neutrophilic (Neutro) Haemophilus (Haemo), Neutro balanced (B), and eosinophilic (Eosino) subgroups (LDA > 4.0, false discovery rate [FDR] P < 0.05). The average relative abundances ranked from high to low are shown for each genus across the three subgroups in the COPDMAP (Chronic Obstructive Pulmonary Disease [COPD] Medical Research Council/Association of the British Pharmaceutical Industry) and AERIS (Acute Exacerbation and Respiratory Infections in COPD) cohorts. (B) Box-and-whisker plots showing microbiome genera most enriched in each subgroup (LDA, FDR P < 0.05). (C) LEfSe analysis showing enrichment (blue) or depletion (red) of the 12 genera in the Eosino versus Neutro group in multiple analyses by 1) comparing the Eosino and Neutro B subgroups and using two additional approaches to control for Haemo overgrowth in the Neutro group, by rescaling relative abundances with Haemo abundance downscaled to its average across samples according to the method used by Taylor and colleagues (21) (Haemo rescaled), and by using a Quantile norm approach to rescale relative abundances to their within-sample percentile ranks; 2) subanalyzing within the initial, second, and third stable visits and during exacerbations; 3) subanalyzing within each of the four sites; and 4) using BEAT-COPD (Biomarkers to Target Antibiotic and Systemic COPD) data. The LDA score for each specific comparison is indicated in the corresponding cell in the table. (D) An example illustrating the time-series analysis on longitudinal microbiome data. Shown are the changes in relative abundances of Campylobacter compared with the changes in Neutro and Eosino status across visits for one patient (South subject-137). The break in between the red lines indicates significant changes in the relative abundance of Campylobacter identified by the changepoint-detection algorithm, which coincided with the switch from the Neutro to the Eosino state. The changes in abundance of Campylobacter were also in concert with sputum Eosino percentages over time, with a cross-covariance (cross-cov) score of 0.861. (E) The microbiome genera whose change points were associated with exacerbation events and with switches between Neutro and Eosino inflammation within stable disease. The odds ratio and 95% confidence interval (95% CI) are shown. Only significant genera with lower limit of the 95% CI greater than 1.0 are shown. (F) The top 10 genera with greatest cumulative cross-cov scores with sputum Eosino percentages. The cumulative cross-cov score and interpatient stdev of the scores were shown for each genus. The genera were highlighted in asterisks if their cross-cov scores were significantly higher than the null distributions derived from permutation test. *FDR P < 0.05. **FDR P < 0.01, and ***FDR P < 0.001. adj = adjusted; H = Haemo-predominant; Leic = Leicester; Manc = Manchester; Quantile norm = quantile normalization; South = Southampton; stdev = SD.
