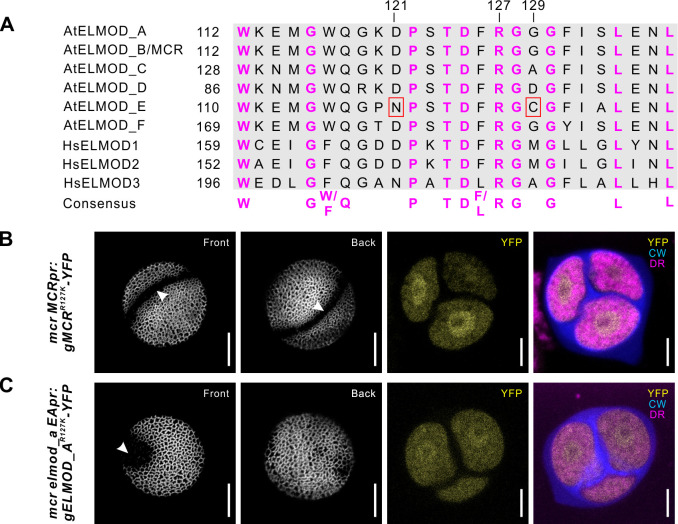
Figure 6. The R127 residue of MCR and ELMOD_A is essential for aperture formation.
(A) Sequence alignment of the conserved GAP regions from six Arabidopsis (At) and three human (Hs) ELMOD proteins, along with the consensus sequence. Invariant Arg residue (R127) and two other important residues (121 and 129) are indicated. N121 and C129, essential for AtELMOD_E function, are indicated by red squares. (B, C) Confocal images of pollen grains and tetrads from mcr and mcr elmod_a expressing, respectively, MCRpr:MCRR127K-YFP (B) and EApr:ELMOD_AR127K-YFP (C). Apertures are indicated with arrowheads. Scale bars, 10 μm for pollen and 5 μm for tetrads.