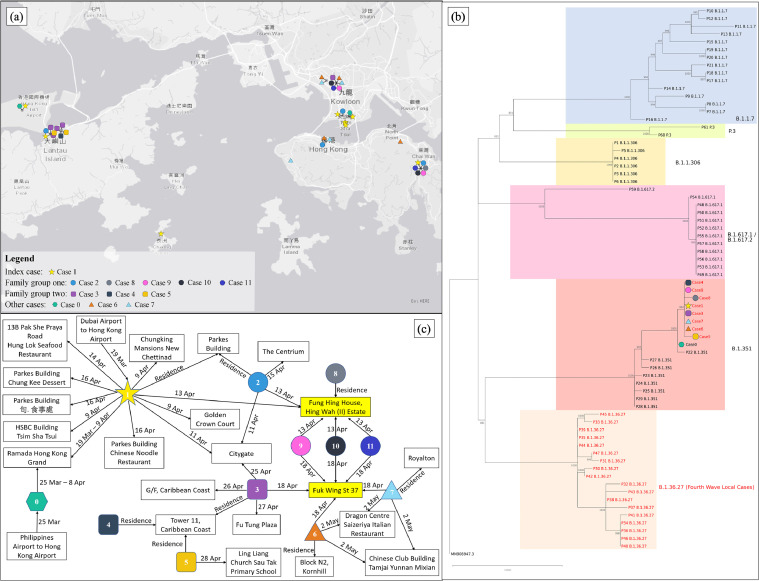
Figure 4.
Phylogeographic analysis of the genomic and spatial relationship among the patients with SARS-CoV-2 VOC B.1.351 in Hong Kong. (a) A geographic map of Hong Kong illustrated the locations of the premises visited by the patients. Cases were linked up with black lines if they visited the same premise on the same day. (b) A phylogenetic tree involved 70 COVID-19 cases identified in April 2021. The tree was rooted on the earliest published genome (accession no.: MN_908947.3). The cases written in red indicated locally-acquired cases, whereas the cases written in black were imported cases. The letters on the right were the PANGO lineages of the cases in the respective colored boxes. The cases of concerns in this study were donated by the same symbols used in (a). Please note that the genomes of cases 2, 10 and 11 were not available as viral loads in their specimens were too low (Ct value >38) to be sequenced. (c) A linkage chart displayed spatial relationships among the patients with SARS-CoV-2 VOC B.1.351. The date on solid line indicated when the patient visited the respective premises. The boxes highlighted in yellow indicated the premises where some of the patients had gathering and direct transmission was believed to occur.