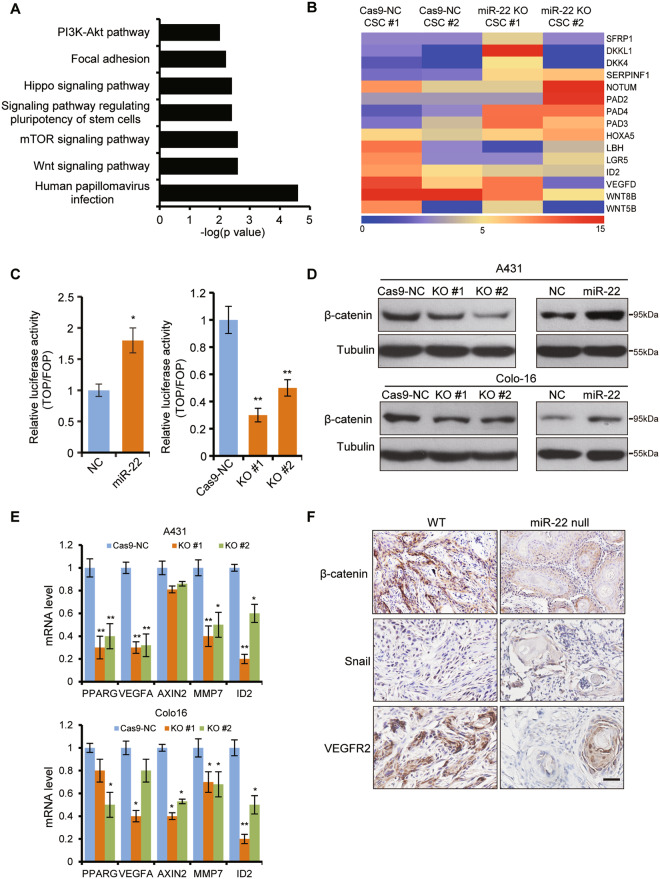
Fig. 5. Wnt/β-catenin signaling was abrogated in miR-22 knockout cells and tissues.
A RNAseq of Cas9-NC (n = 2) and miR-22 KO (n = 2) cells spheroids was performed and KEGG pathway enrichment of the downregulated genes was analyzed. B Heatmap for Wnt/β-catenin pathway inhibitors (SFRP1, DKKL1, DKK4, SERPINF1, NOTUM, PAD2, PAD4, PAD3, HOXA5) and effectors (LBH, LGR5, ID2, VEGFD, WNT8B, WNT5B). This heatmap was generated and analyzed based on the fold change of the differentially expressed genes between miR-22 knockout and wild-type cSCC cell spheroids. C, D TOPflash reporter activity, and β-catenin protein level was determined in miR-22 overexpressed cells (miR-22) and its negative control cells (NC), Cas9-NC cells, and miR-22 KO cells. n = 3 biological replicates. *p < 0.05, **p < 0.01. E The mRNA level of Wnt/β-catenin related downstream factors were analyzed by qPCR in Cas9-NC cells and miR-22 KO cells. *p < 0.05, **p < 0.01. n = 3 biological replicates. F Immunohistochemistry (IHC) analysis for β-catenin, Snail, and VEGFR2 in cSCC from WT (n = 3) and miR-22 null mice (n = 3). Scale bar, 50 µm.