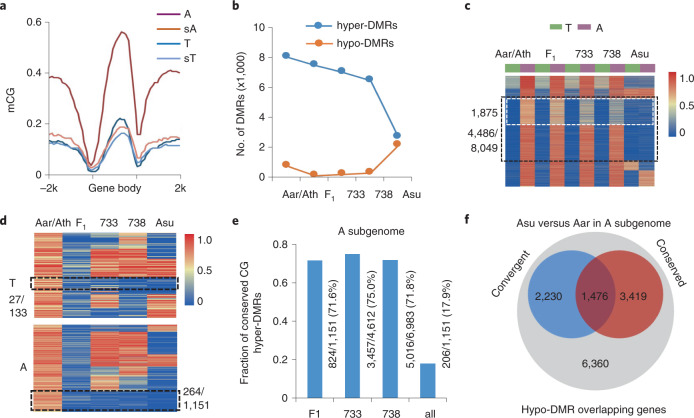
Fig. 4. Convergence and inheritance of CG methylation levels between two subgenomes in allotetraploids.
a, CG methylation levels of homologues in A. thaliana (Ler4, T), A. arenosa (Aar, A), sT and sA subgenomes of A. suecica. b, Numbers of DMRs between T subgenome and Ath (Col) or A subgenome and Aar in F1, 733, 738 and Asu, respectively. Note that Allo733 and Allo738 may be treated as biological replicates of resynthesized allotetraploids. c, Clustering analysis of CG hyper-DMRs (A-T) in Aar/Ath and their changes in F1, Allo733 (733), Allo738 (738) and A. suecica (Asu), respectively. Dashed black box indicates 4,486 convergent DMRs where hyper-DMRs between A and T (Ler4) were conserved in newly formed allotetraploids and reduced to the sT subgenome level in Asu. Note that the upper portion (white dashed box) indicates the overlap group (1,875) with conserved DMRs (also see e). d, Clustering of CG hypo-DMRs in F1 and their changes in 733, 738 and Asu relative to Aar/Ath. Black dashed boxes indicate hypo-DMRs between T subgenome and Ath (upper panel) and between A subgenome and Aar (lower panel) in F1 were conserved in Allo733, Allo738 and Asu. e, Fraction of conserved CG hypo-DMRs in F1, Allo733, Allo738 and all three lines and their inheritance in Asu relative to Aar, with the numbers (conserved/total) shown to the left of each column. f, Venn diagram of the genes that overlapped with convergent (blue) and conserved (red) CG hypo-DMRs in Asu relative to Aar. Absolute values of CG methylation change thresholds were 0.5 in c,d.