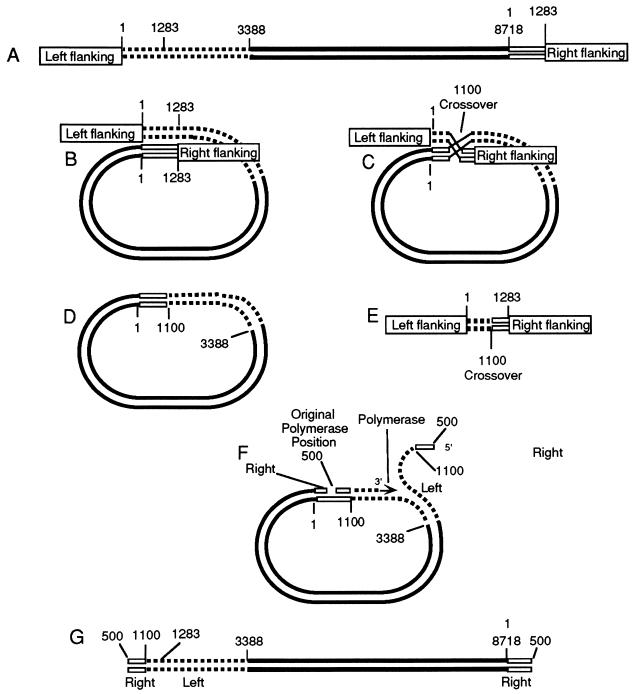
FIG. 10.
Model for the formation of macronuclear DNA from micronuclear DNA. (A) Linear micronuclear RIB2. (B) The molecule bends back on itself, and homologous regions pair. (C) Next there is a crossover at about base 1100, which cuts out the region with the deletion of the micronuclear gene. (D) Resultant circular form of the micronuclear gene. (E) Defective region that was cut out. (F) A polymerase molecule now attaches at about base 500 and begins to transcribe in a typical rolling-circle replication. The left part of the molecule peeling off the template begins at about base 500, and the polymerase continues to transcribe, going around the circle to the position where replication started. (G) Molecule produced by one cycle of replication. The polymerase continues around the circle numerous times to produce a continuous sequence of transcribed and nontranscribed regions. Finally, replication stops near the point where it started, producing the right end of the molecule. After a complementary strand has been synthesized, telomeres are then added to the free ends of the linear molecule to produce the macronuclear polymer of RIBX. Alternatively, the two free ends, instead of adding telomeres, may combine to form a circle.