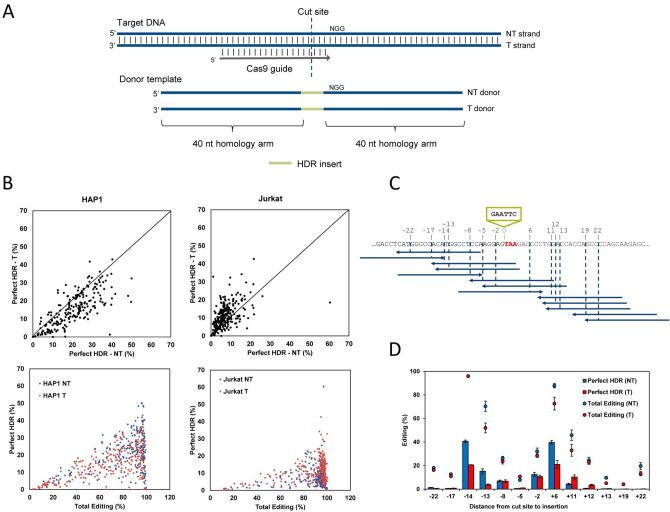
Figure 1.
Cas9 HDR strand preference and gRNA selection. (A) Schematic representation of targeting (T) and non-targeting (NT) donor template designs. The targeting strand is complementary to the gRNA sequence, whereas the non-targeting strand contains the guide and PAM sequence. (B) An EcoRI recognition site was inserted at a Cas9 cleavage site at 254 genomic loci in Jurkat and 239 genomic loci in HAP1 cells using either the T or NT strand as the donor template. RNP complexes (Alt-R S.p. Cas9 Nuclease complexed with Alt-R CRISPR–Cas9 crRNA and tracrRNA) were delivered at 4 µM along with 4 µM Alt-R Cas9 Electroporation Enhancer and 3 µM donor template by nucleofection. Total editing and perfect HDR was assessed via NGS. (C) Schematic of the gRNAs used to facilitate HDR insertion of an EcoRI site before the stop codon of GAPDH (TAA, red) in K562 cells using 13 guides around the desired HDR insertion location (blue, arrows indicate the 3’ end). The cleavage sites and associated distance to the desired insertion location (green) for each gRNA are indicated above the sequence shown. Both the T and NT strand were tested. (D) RNP complexes (Alt-R S.p. Cas9 Nuclease, Alt-R CRISPR–Cas9 crRNA and tracrRNA) for the 13 guides targeting GAPDH were delivered at 2 µM along with 2 µM Alt-R Cas9 Electroporation Enhancer and 2 µM donor template designed to insert an EcoRI site before the stop codon by nucleofection to K562 cells. HDR and total editing were assessed via NGS. Data are represented as means ± SEM of three biological replicates.