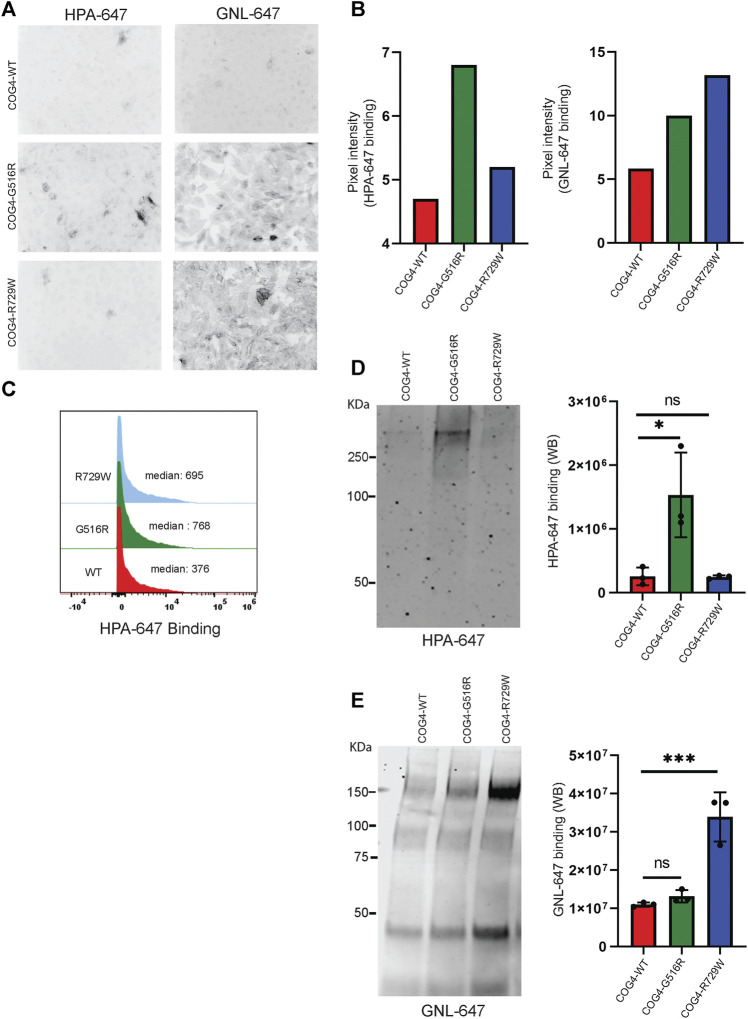
FIGURE 6.
COG4-G516R and COG4-R729W mutations are causing O-glycosylation and N-glycosylation defects, respectively. (A) Non-permeabilized RPE1 rescued COG4-WT-3myc, COG4-G516R-3myc, and COG4-R729W-3myc cells were stained with the fluorescently conjugated lectins HPA-647 (specific for terminal GalNAc residues) and GNL-647 (specific for terminal α-D-mannosyl residues). The images were taken by Zeiss Axiovert 200M fluorescent microscope using a ×20 objective. (B) The pixel intensity of the full image for each cell line has been measured using ImageJ software. The bar graph has been presented with statistical value to provide intensity difference for each lectin (HPA-647, GNL-647) binding among the rescued cell line. (C) Flow cytometry histogram of HPA-647 fluorescence intensity in COG4-WT-3myc (red), COG4-G516R-3myc (green), and COG4-R729W-3myc (blue) population after gating for live and single cells. (D) Western blot analysis of the secretion from the rescued cells has been performed by probing with the HPA-647 antibody (left panel). Quantification of the lectin HPA-647 binding intensity (Right panel). Statistical significance was calculated by GraphPad Prism 8 using One-way Anova. *p ≤ 0.05, **p ≤ 0.01 and non-significant (ns) p > 0.05. Error bar represents mean ± SD. (E) Western blot analysis of the secretion from the rescued cells has been performed by probing with the GNL-647 antibody (left panel). Quantification of the lectin GNL-647 binding intensity (Right panel). Statistical significance was calculated by GraphPad Prism 8 using One-way ANOVA. *p ≤ 0.05, **p ≤ 0.01 and non-significant (ns) p > 0.05. Error bar represents mean ± SD.