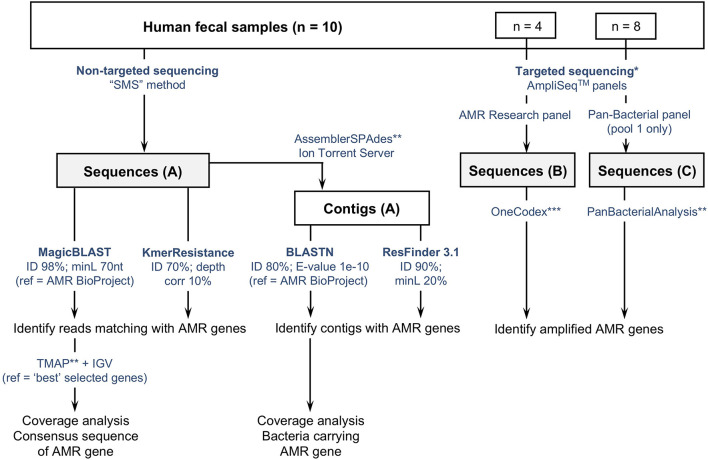
FIGURE 1.
Flowchart describing three culture-independent methods to identify the presence of antimicrobial resistance genes (ARGs) in fecal samples (n = 10). (A) Non-targeted sequencing of sheared DNA or “Shotgun Metagenomic Sequencing” (SMS) method; (B,C) targeted sequencing using two commercially available AmpliSeqTM panels targeting ARGs. Sequences obtained with the SMS method were analyzed using two web-based tools (ResFinder and KmerResistance, https://cge.cbs.dtu.dk/services/), and a method based on the comparison of reads and contigs against the NCBI-curated “Bacterial Antimicrobial Resistance Reference Gene Database” (https://www.ncbi.nlm.nih.gov/bioproject/PRJNA313047). *Targeted sequencing was tested on a subset of samples: 4 with the AMR Research panel, 8 with the Pan-Bacterial Research panel. **refers to Ion Torrent Suite plugins available from the Thermo Fisher Scientific Plugin store. ***Refers to the “One Codex” online platform (available at: https://app.onecodex.com/) used with the AMR Research panel. “ID”: percentage identity threshold; “minL”: minimum length: minimum percentage of coverage compared to reference length or minimum number of nucleotides (nt); “ref”: reference sequences used for comparison with reads and/or contigs.