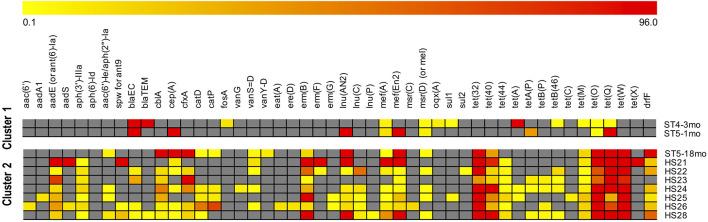
FIGURE 4.
Clustering of samples by antimicrobial resistances genes (ARGs) abundance. ARGs were identified with the SMS + BLAST analysis. The samples identifiers are indicated on the right side (ST are infants, HS are adults). The “mean base depth per 5 million reads” (BD5M) calculated for each ARG in each sample was used as an input for the k-means clustering (1,000 maximum iterations, 50 runs). The color scale is: gray when the gene is absent; from light yellow to dark red with increasing gene abundances (BD5M 0.1–96), with values adjusted to log2. To avoid one outlier effect [gene tet(A) in ST4-3mo; BD5M = 279.8] the maximum value was set as the second highest abundance [gene tet(X) in HS24; BD5M = 96.2].