Figure 3.
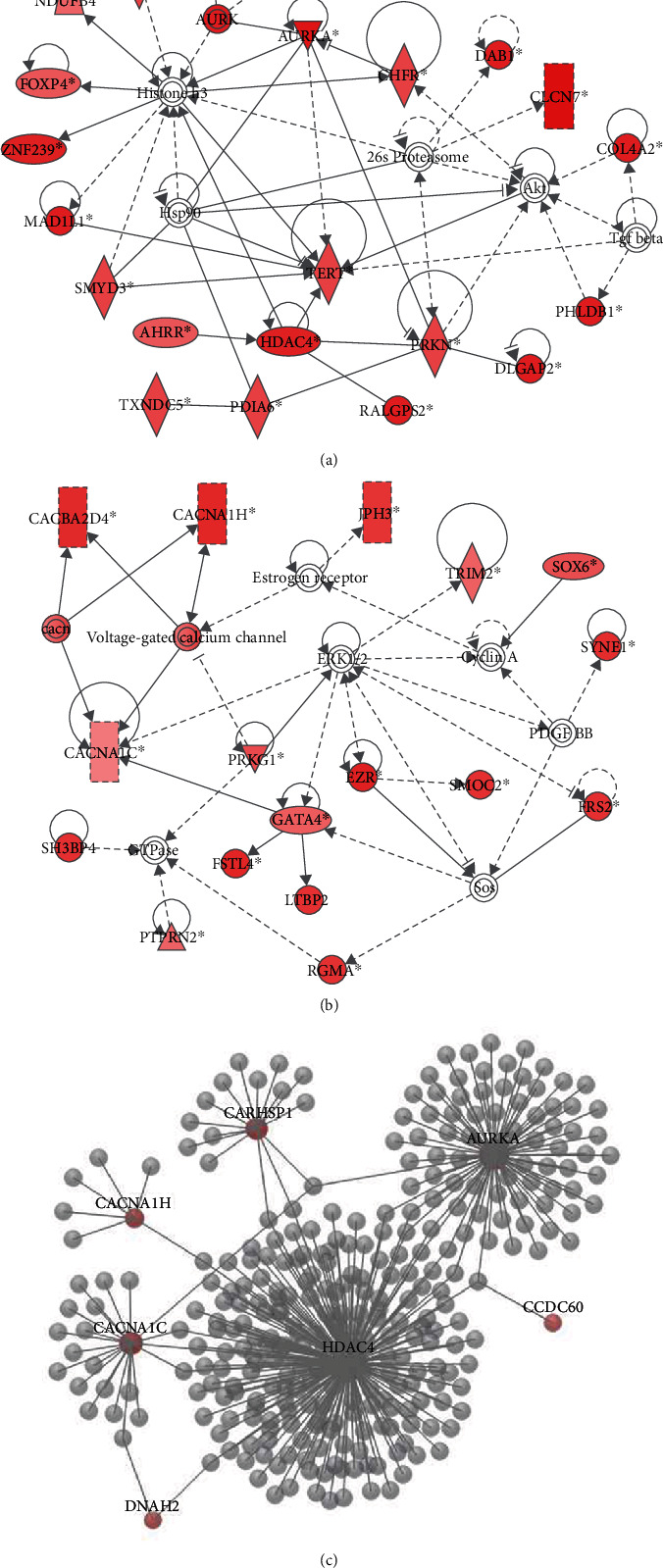
Top-ranked functional networks of the differentially methylated genes. (a) Top network identified by ingenuity pathway analysis (IPA) of differentially methylated genes related to cell cycle, cellular assembly, and organization. (b) Top network identified by IPA of differentially methylated genes related to organ development and function. Colored nodes indicate differentially methylated genes. Noncolored nodes were proposed by IPA and suggest potential targets functionally coordinated with the differentially methylated genes. Dashed lines represent indirect relationships, and solid lines indicate direct molecular interactions. In each network, edge types are indicatives: a line without arrowhead indicates binding only; a line finishing with a vertical line indicates inhibition; a line with an arrowhead indicates ‘acts on.' (c) Protein-protein interaction network of selected differentially methylated genes. Using the OmicsNet database, six genes (AURKA, HDAC4, CARHSP1, CACNA1H, CACNA1C, and DNAH2) were used to construct a top-ranked functional protein-protein interaction network.
