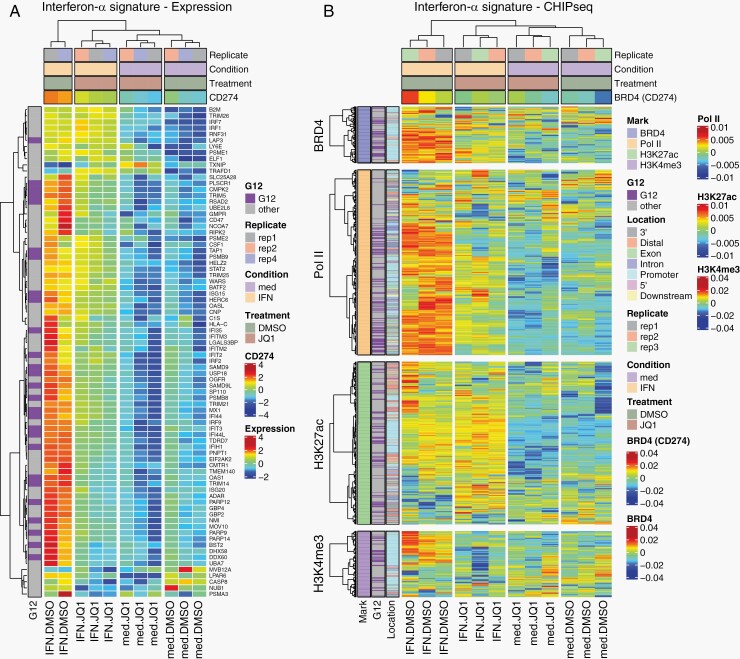
Fig. 4.
JQ1 impairs the transcriptional program activated by IFN-α. (A) Heatmap representation of RNA-seq for ISGs from LN-2683GS under 4 experimental conditions: LN-2683GS cells were treated for 2 hours with IFN-α, 1 µM JQ1, or the combination thereof. The samples are annotated for treatment (JQ1, DMSO), condition (IFN-α, media [med]), and replicate. The normalized CD274 expression was added as a supplementary variable. Genes overlapping with the G12 GBM IFN-signature are annotated in a separate column (G12, purple, nonG12, gray). (B) Heatmap shows consensus peak intensities (ChIP-seq) of BRD4, Pol II, H3K27ac, and H3K4me3 at ISGs in LN-2683GS under 4 experimental conditions as in (A). The sample dendrogram is based on the BRD4 ChIP-seq dataset. The BRD4 consensus peak of CD274 was added as a supplementary variable. The consensus peak locations (promoter, intergenic, exon, intron, etc.) of BRD4, Pol II, H3K27ac, and H3K4me3 are annotated in a color code as indicated. The peaks overlapping with G12 genes are annotated (G12, purple; nonG12, gray). Abbreviations: DMSO, dimethyl sulfoxide; GBM, glioblastoma; IFN-α, interferon alpha; ISGs, interferon-stimulated genes.