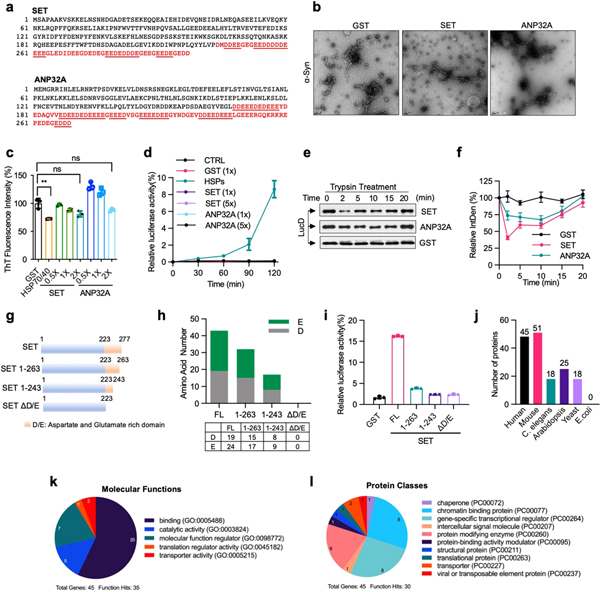
Extended Data Fig. 6 |. Role of other polyD/E proteins in protein folding.
a, Sequences of human SET and ANP32A proteins. The poly D/E region is marked in red colour, and the continuous runs of Asp/Glu are underlined. b, c, ANP32A and SET does not block α-Syn fibrillization. α-Syn monomers (70 μM) were incubated with SET or ANP32A (0.4 μM each) for 7 days. Samples were analysed by electron microscopy (b) and ThT staining (c). Scale bar, 100 nm. d, SET and ANP32A are unable to solubilize urea-denatured luciferase. Urea-denatured luciferase (5 nM) was incubated with GST, SET, ANP32A (0.2 μM each) or HSPs (0.2 μM HSP70, 0.1 μM HSP40, and 0.4 μM HSP104(A503S)) at 25 °C for 90 min. Activity relative to that of the native control are shown. e, f, Unfoldase activity of ANP32A and SET. Misfolded LucD (50 nM) was incubated with GST, SET or ANP32A (200 nM each) at 25 °C. At indicated time points, aliquots of refolding luciferase were incubated with 2.5 μM trypsin at 22 °C for 2 min, denatured in SDS sample buffer, and analysed by western blot (e), with the quantification showed in (f). g, h, Schematic presentation of SET and its deletion mutants (g), and the numbers of Asp (D) and Glu (E) in each mutant (h). i, Heat-inactivated luciferase (5 nM) was incubated at 25 °C with SET or its deletion mutants (200 nM each) for 90 min. Activity relative to that of native luciferase are shown. j, Number of polyD/E proteins different species. k, l, Gene Ontology analysis of polyD/E proteins in humans. Proteins are classified into pie chart based on their molecular functions (k) and protein classes (l). Assays have been performed two (b) or three (e) times with similar results. Numerical data are mean ± s.d. (n = 3) and are representative of two independent experiments (c, d, f, i). **P < 0.01, unpaired Student’s t-test.