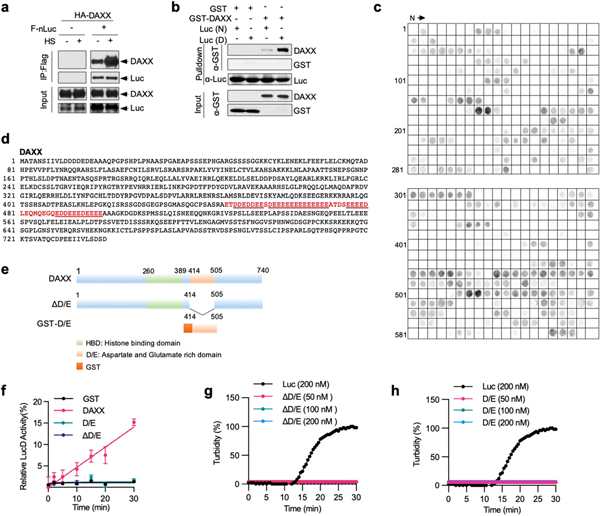
Extended Data Fig. 5 |. DAXX binds to misfolded proteins and depends on the polyD/E region for its activity.
a, The DAXX-luciferase interaction is increased upon heat shock. HEK293T cells were transfected with HA–DAXX and Flag–nLuc as indicated, and treated with or without heat shock. Cell lysates were immunoprecipitated with anti-Flag mAb (M2) beads. Input and immunoprecipitated samples were analysed by western blot. b, DAXX preferentially binds to heat-denatured luciferase. GST or GST–DAXX (100 nM each) was incubated with native (N) or heat-denatured (D) 6xHis-luciferase immobilized on Ni-NTA agarose. The input and pulldown samples were analysed by western blot. c, Binding of DAXX to cellulose-bound peptide scans derived from luciferase, p53, MDM2, H3.3, H4 and DAXX. Each peptide contained thirteen amino acid residues that overlapped adjacent peptides by ten. d, Protein sequence of DAXX. The poly D/E region is marked in red colour, and the four continuous runs of Asp/Glu (with 14, 11, 7, and 5 residues, respectively) are underlined. e, Schematic presentation of full-length DAXX and its mutants. DAXX(D/E) was fused to GST at the N terminus for protein stabilization. f, DAXX(ΔD/E) and DAXX(D/E) lack unfoldase activity. Misfolded LucD monomers (50 nM) were incubated with DAXX(ΔD/E) or DAXX(D/) (100 nM each) for the indicated times and assayed for luciferase activity (,ean ± s.d., n = 3). g, h, DAXX(ΔD/E) and DAXX(D/E) remain soluble during heat treatment. Recombinant DAXX ΔD/E (g) and D/E (h) proteins were heated at 42 °C for the indicated durations. Luciferase (200 nM) was used as a positive control. Aggregation formation was monitored by OD600 and normalized to the luciferase control. Assays in a–c have been performed two times with similar results. Numerical data are mean or mean ± s.d. (n = 3) and are representative of two independent experiments (f–h).