Figure 4. Purine synthesis and its dependency on mitochondrial metabolism.
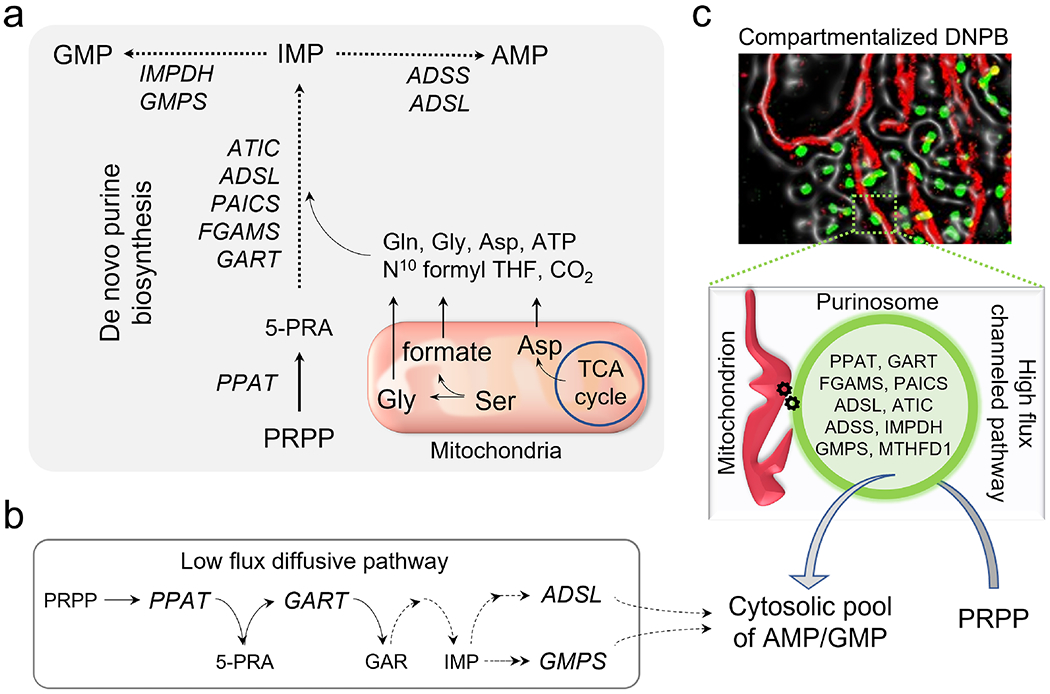
(a)De novo purine biosynthesis (DNPB) converts phosphoribosyl pyrophosphate (PRPP) to inosine monophosphate (IMP) which is bifurcated into the products adenosine monophosphate (AMP) and guanosine monophosphate (GMP) employing nine enzymes: amidophosphoribosyl transferase (PPAT), trifunctional phosphoribosylglycinamide formyltransferase (GART), phoshoribosylformylglycinimidine transferase (PFAS/ FGAMS), bifunctional phosphoribosylaminoimidazole carboxylase and phosphoribosyl aminoimidazole succinocarboxamide synthase (PAICS), bifunctional adenylosuccinate lyase (ADSL), bifunctional 5-aminoimidazole-4-carboxamide nucleotide formyltransferase/ IMP cyclohydroxylase (ATIC), adenylosuccinate synthetase (ADSS), IMP dehydrogenase (IMPDH), GMP synthetase (GMPS). Generation of the pathway substrates glycine (Gly), aspartic acid (Asp), and cofactor N 10formyl-tetrahydrofolate (N10 formyl THF) depends on mitochondrial metabolism. Solid arrows: single reaction step, dashed arrows: multiple steps in the cascade. (b) Under purine depletion, HeLa cells perform diffusive DNPB with only a low flux contribution. Abbreviations: 5-phosphoribosylamine (5-PRA) and glycinamide ribonucleotide (GAR). (c) Channeled synthesis utilizing mitochondria (labeled in red) associated purinosomes (labeled in green) visualized by fluorescence imaging in HeLa cells is the major contributor to the overall DNPB flux. Note the inclusion in the purinosome of methylenetetrahydrofolate dehydrogenase/cyclohydrolase (cytosolic isoform MTHFD1). The factors responsible for stabilizing purinosomes on mitochondrial membranes are not yet known.
