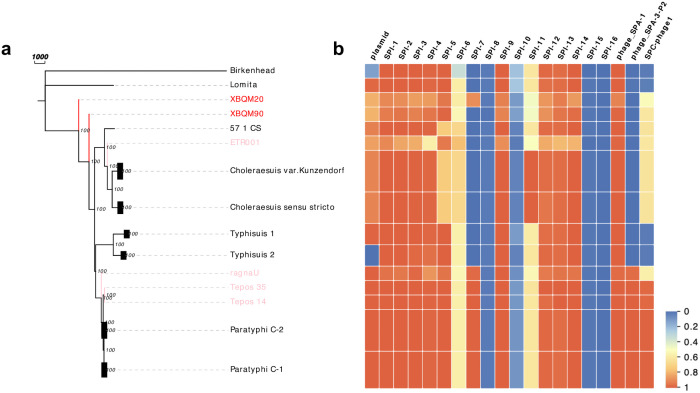
Fig 4. Phylogenetic tree of the Para C lineage and the gain and loss of virulence factors.
A. Maximum parsimony tree of the Para C lineage. Paratyphi C, Typhisuis and Choleraesuis strains are collapsed based on their predicted serovar in order to better represent the relevant information and the Birkenhead serovar was used as the outgroup. B. Covered percentage of Salmonella virulence factors including the pSPCV plasmid, Salmonella pathogenicity islands (SPI1-SPI16) and prophages. For grouped strains, such as Paratyphi, Typhisuis and Choleraesuis, we showed the average percentage of covered region.