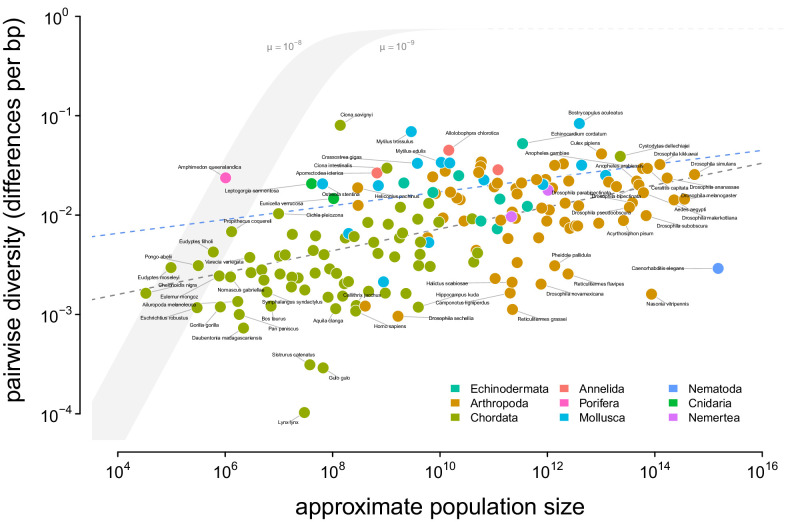
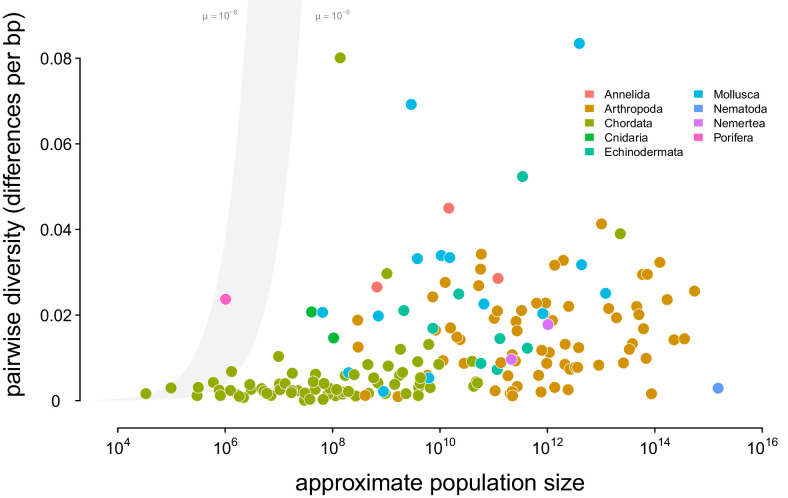
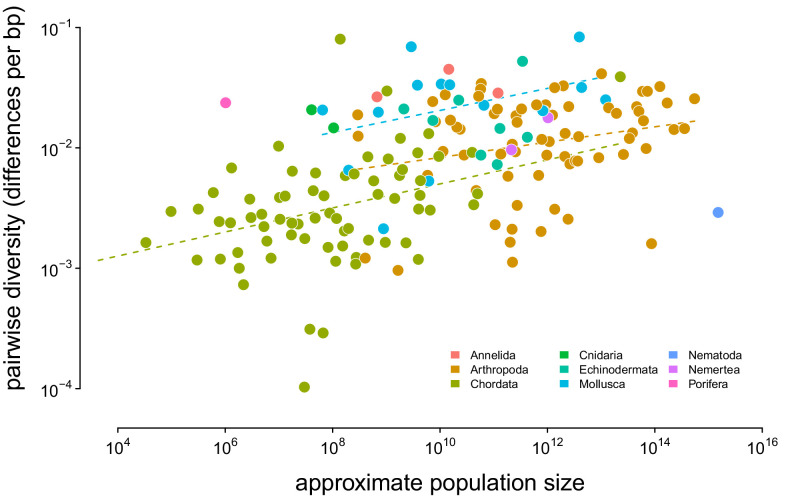
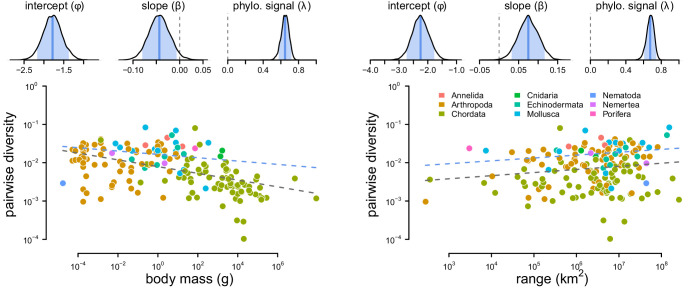
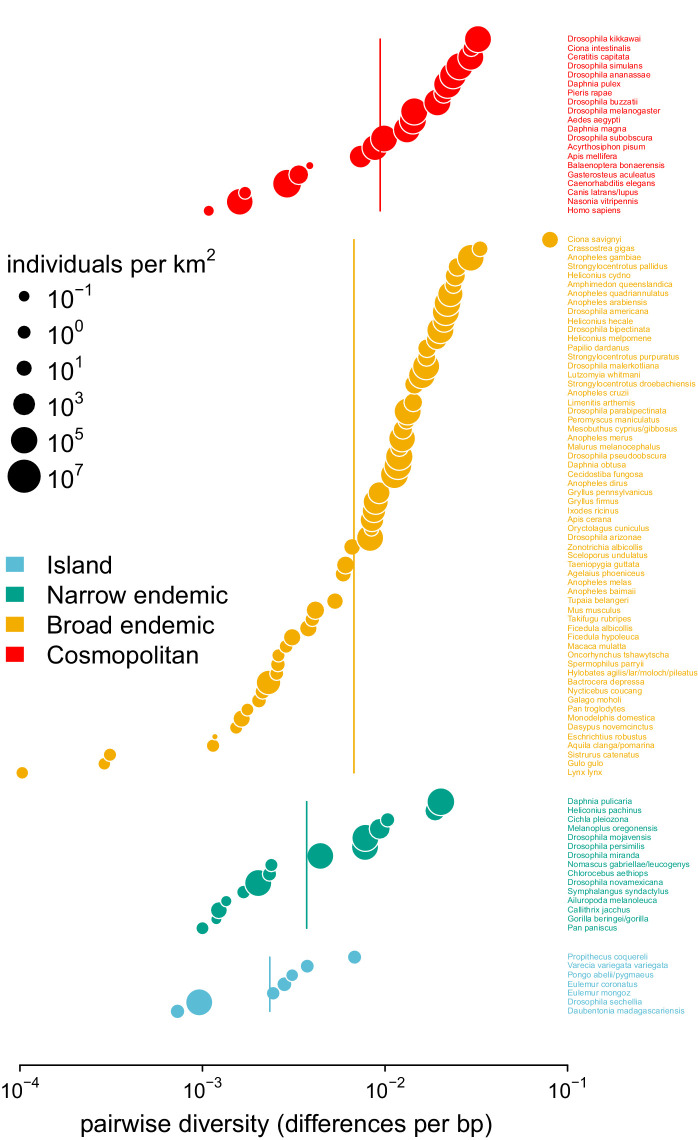
Figure 2. A visualization of Lewontin’s Paradox of Variation.
Pairwise diversity (data from Leffler et al., 2012, Corbett-Detig et al., 2015, and Romiguier et al., 2014), which varies over three orders of magnitude, shows a weak relationship with approximate population size, which varies over 12 orders of magnitude. The shaded curve shows the range of expected neutral diversity if were to equal under the four-alleles model, where , for two mutation rates, and , and the light gray dashed line represents the maximum pairwise diversity under the four alleles model. The dark gray dashed line is the OLS regression fit, and the blue dashed line is the regression fit using a phylogenetic mixed-effects model. Points are colored by phylum. The species Equus ferus przewalskii ( and ) was an outlier and excluded from this figure for visual clarity.