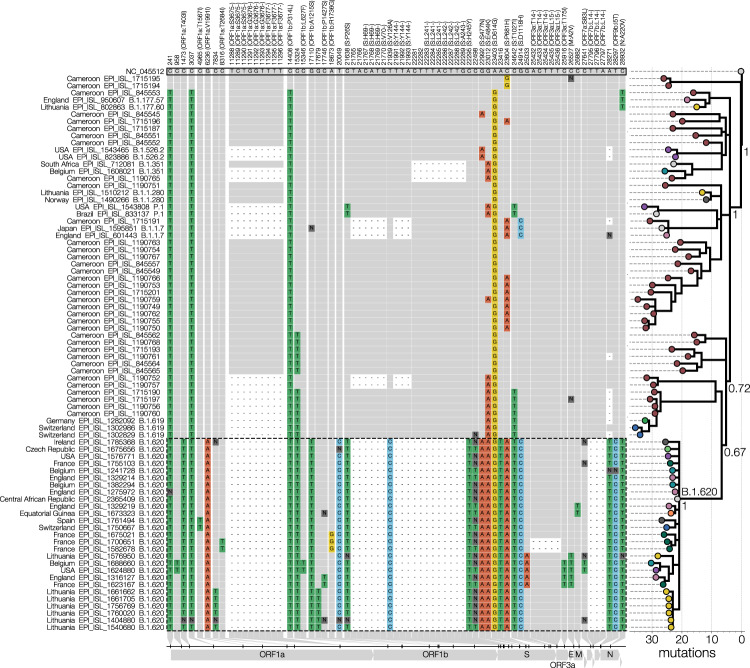
Fig. 1. Lineage-defining SNPs of lineage B.1.620.
Only SNPs that differentiate B.1.620 (genomes outlined with a dashed line) from the reference (GenBank accession NC_045512) and that are shared by at least two B.1.620 genomes are shown in the condensed SNP alignment. Sites identical to the reference are shown in grey, changes from the reference are indicated and coloured by nucleotide (green for thymidine, red for adenosine, blue for cytosine, yellow for guanine, dark grey for ambiguities, black for gaps). The first 100 and the last 50 nucleotides are not included in the figure but were used to infer the phylogeny. If a mutation results in an amino acid change, the column label indicates the gene, reference amino acid, amino acid site, and amino acid change in brackets. The phylogeny (branch lengths in the number of mutations) on the right shows the relationships between depicted genomes and was rooted on the reference sequence with coloured circles at the tips indicating the country from which the genome came. Posterior probabilities of nodes leading up to lineage B.1.620 are shown near each node with the long branch leading to lineage B.1.620 labelled as ‘B.1.620’.