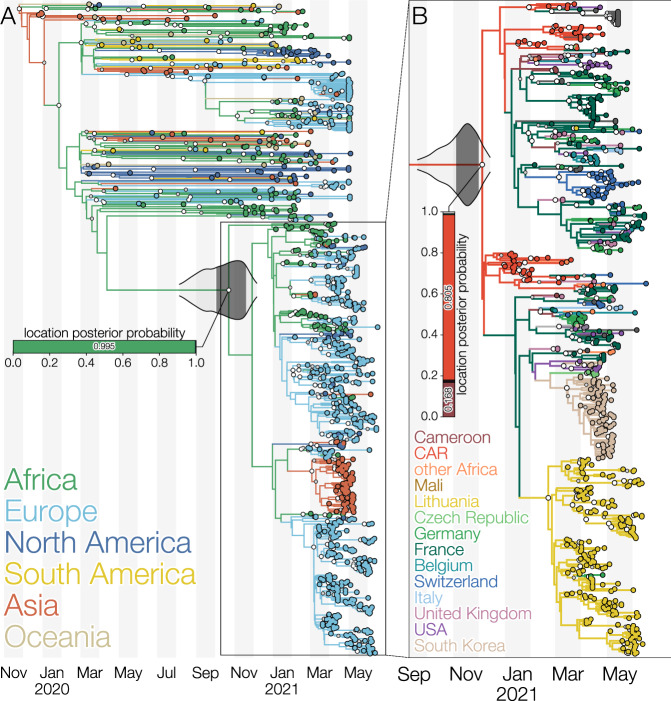
Fig. 4. Maximum clade credibility trees of lineage B.1.620 coloured by reconstructed location using the latest available data as of June 2021.
A Global phylogeny of SARS-CoV-2 genomes with branches coloured by inferred continent from a Bayesian phylogeographic analysis that makes use of individual travel histories. Lineage B.1.620 is outlined and a horizontal bar shows the posterior probability of its common ancestor existing in a given continent. Africa is reconstructed as the most likely location (posterior probability 0.995) where B.1.620 originated. The 95% and 50% highest posterior density (HPD) intervals for the most recent common ancestor date of lineage B.1.620 are indicated with violin plots centred on the common ancestor. B Phylogeny of lineage B.1.620 with branches coloured by inferred country from a Bayesian phylogeographic analysis that makes use of travel histories. A vertical bar shows posterior probabilities of where the common ancestor of B.1.620 existed. In this analysis, Central African Republic (CAR) and Cameroon are reconstructed as the most likely locations (with posterior probabilities of 0.805 and 0.168, respectively) of the common ancestor of lineage B.1.620. Larger white dots at nodes indicate nodes with a posterior probability of at least 95%, while smaller grey circles indicate nodes with a posterior probability of at least 50%. The 95% and 50% highest posterior density (HPD) intervals for the most recent common ancestor date of lineage B.1.620 are indicated with violin plots centred on the common ancestor.