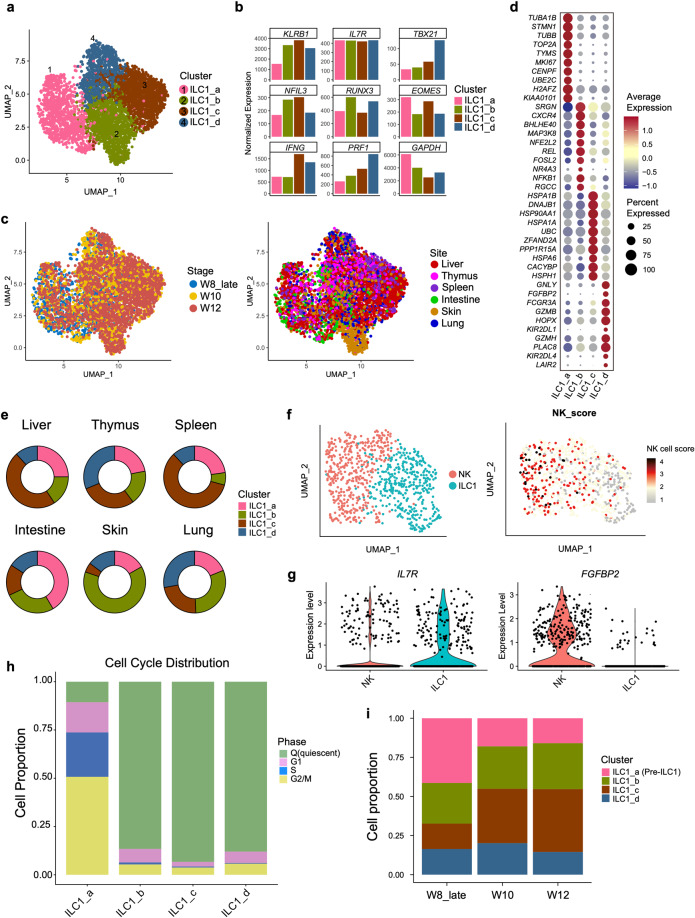
Fig. 4. Heterogeneity and tissue distribution bias of Group 1 ILCs.
a UMAP visualization of Group 1 ILCs from Fig. 2a with re-clustered populations mapped on. Colors indicate cell types. b Expression of the known ILC1-related genes in the different ILC1 sub-clusters is shown in bar graph. c Stage (left panel) and site (right panel) information of ILC1 sub-clusters identified in a, indicated by colors. d Dot plots show top 10 DEGs in the ILC1_a, ILC1_b, ILC1_c, ILC1_d clusters. Colors represent gene expression levels, and size encodes the proportion of gene-expressing cells. e Pie charts show the proportions of ILC1_a, ILC1_b, ILC1_c and ILC1_d in tissues. Colors represent cell clusters described in a. f UMAP visualization of the cells in ILC1_d cluster are further separated into NK and ILC1 subgroup by UMAP analysis (left panel). NK score (GEO: GSE70580) is projected onto UMAP visualization, and colors indicate the expression level of NK-related genes (right panel). g Violin plots show the expression level of IL7R and FGFBP2 in NK and ILC1 subgroups defined in f. h The proportions of cell in different cell cycle statues in ILC1 sub-clusters are shown in bar graph. Colors indicate different cell cycle statuses. i The proportions of ILC1 sub-clusters in each embryonic development stages are shown in bar graph. Colors indicate cell types.