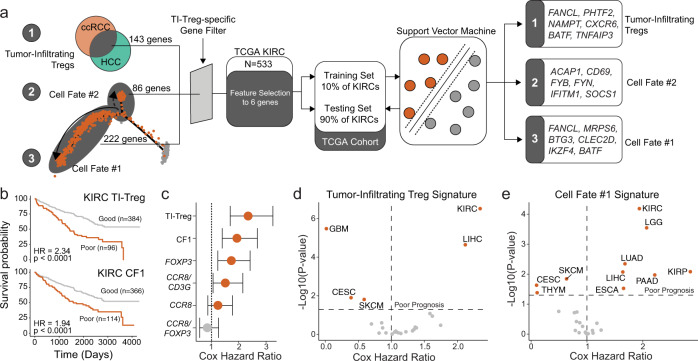
Fig. 3. Improved prognostic prediction is associated with a signature from the suppressive TI Treg cell fate.
a Schematic of signature development using feature selection from: (1) 143 common differential genes of TI Treg in ccRCC and HCC, (2) 86 genes differentially expressed in CF2, and (3) 222 genes differentially expressed in CF1 using the 10% of the TCGA KIRC/ccRCC dataset for feature selection. Gene signatures generated after feature selection were used to predict prognosis in the remaining 90% TCGA KIRC/ccRCC, as well as 23 other TCGA cancer datasets. b Kaplan–Meier curves for overall survival in TCGA KIRC/ccRCC using the TI-Treg common gene signature (upper panel) and CF1-Treg gene signature (lower panel). c Prognostic prediction for Treg signatures compared to other proposed signatures for TI Treg cells. Hazard ratios with the bars representing the 95% confidence intervals, and P values derived from Cox proportional hazard regression modeling. d, e Overall survival prediction with Cox proportional hazard ratio and −log10(P value) based on two-sided log-rank testing across the 24 largest TCGA datasets using d the TI Treg signature and e the CF1 signature. CESC, Cervical squamous cell carcinoma and endocervical adenocarcinoma; ESCA, esophageal carcinoma; GBM, glioblastoma multiforme; KIRC, kidney renal clear cell carcinoma, KIRP, kidney renal papillary cell carcinoma; LGG, low-grade glioma; LIHC, liver hepatocellular carcinoma; LUAD, lung adenocarcinoma; PAAD, pancreatic adenocarcinoma; SKCM, skin-cutaneous melanoma; THYM, thymoma.