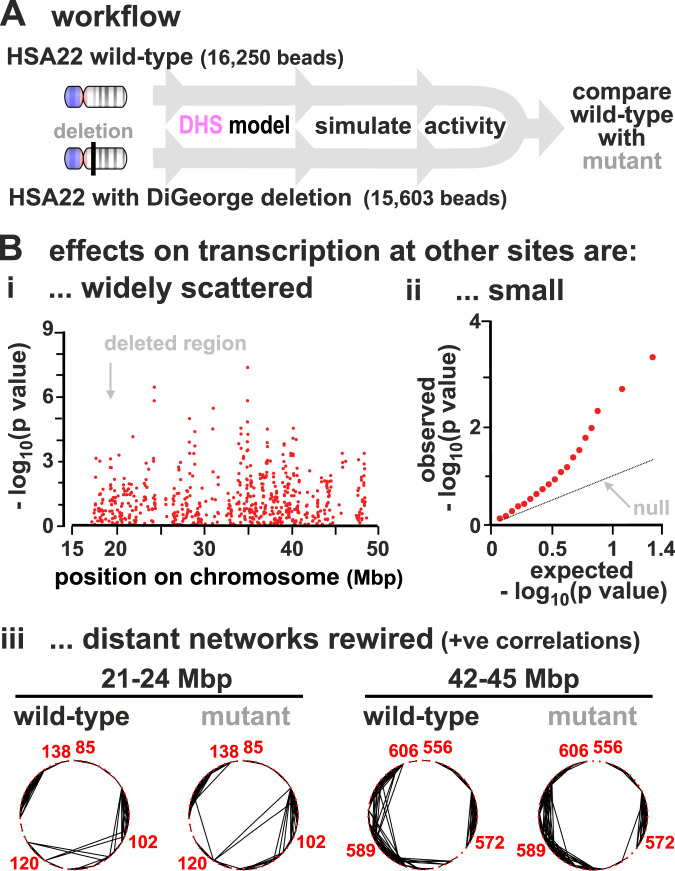
Fig. 8. Modelling effects of the DiGeorge deletion in HSA22.
A Workflow. Simulations (800 simulations/condition) for wild type (17,102 beads) and deletion (16,250 beads, where wild-type beads 6305–7156 are cut, corresponding to a deletion of chr22:18,912,231–21,465,672 in hg19). [Agreement between predicted transcriptional activity and GRO-seq in HSA22 is similar to that found for HSA14 (here, Spearman correlation is r ~ 0.29, p < 10−6; two-sided Student’s t-test).] B (i) Manhattan plot showing (p value) as a function of genomic position along HSA22 (position given in Mbp), for changes in TU transcriptional activities between wild type and deletion. (ii) Quantile–quantile plot showing expected versus observed values for (p value) for the same data in (i). Expected values are computed from the normal distribution (these correspond to the null hypothesis according to which the change in transcriptional activities in the deletion is purely due to random variation). (iii) Regulatory networks of two 3 Mbp segments in chromosome 22 inferred from the Pearson correlation matrix. Edges show positive correlations >0.12 (p = 0.0007). Segments chosen have roughly the same number of nodes in 3 Mbp as the short fragment (Fig. 3Aii).