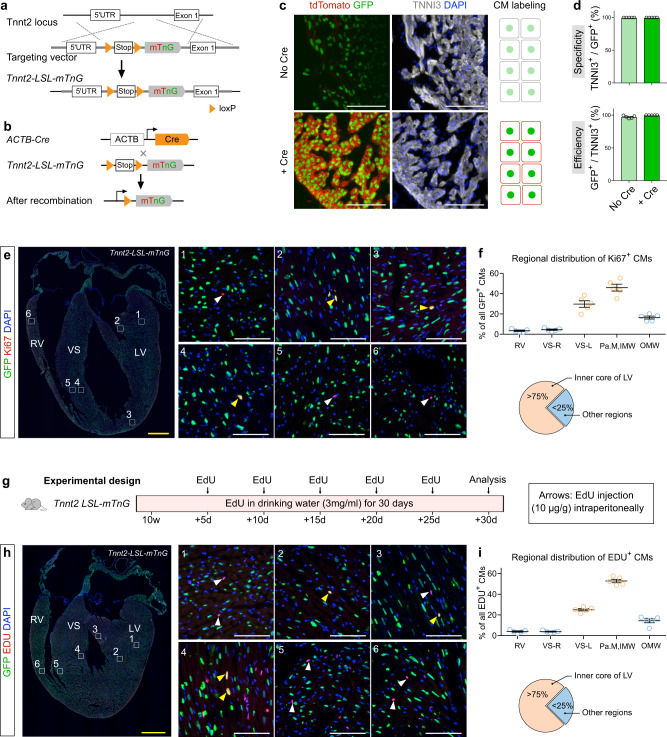
Fig. 6. Cardiomyocyte-specific Ki67 protein expression and EdU incorporation are more enriched in inner core of left ventricle.
a Schematic figure showing generation of Tnnt2-loxP-Stop-loxP-membrane tdTomato-2A-nuclear GFP (Tnnt2-LSL-mTnG) mouse line. b Strategy of crossing Cre line with Tnnt2-LSL-mTnG line. c Immunostaining for GFP, tdTomato, and TNNI3 on heart tissue sections of E13.5 Tnnt2-LSL-mTnG embryos containing Cre or no Cre. Cartoon shows labeling of cardiomyocytes (CM) in no Cre or Cre embryos. d Quantification of the GFP labeling specificity and efficiency in Tnnt2-LSL-mTnG containing Cre or no Cre embryos. Data are mean ± s.e.m.; n = 5. e Immunostaining for Ki67 and GFP on Tnnt2-LSL-mTnG heart sections (10 weeks). Boxed regions (1–6) are magnified on the right panel. Yellow arrowheads, nGFP+Ki67+ CMs; white arrowheads, nGFP–Ki67+ nuclei. f Quantification of the distribution of nGFP+Ki67+ CMs in different regions of hearts. Data are mean ± s.e.m.; n = 5. g Schematic figure showing the experimental design. h Immunostaining for GFP and EdU on heart sections (14 weeks). Boxed regions (1–6) are magnified on the right panel. Yellow arrowheads, nGFP+EdU+ CMs; white arrowheads, nGFP–EdU+ nuclei. i Quantification of the distribution of nGFP+EdU+ CMs in different regions of hearts. Data are mean ± s.e.m.; n = 5. Each image is representative of five individual biological samples. Scale bars: yellow, 1 mm; white, 100 µm.