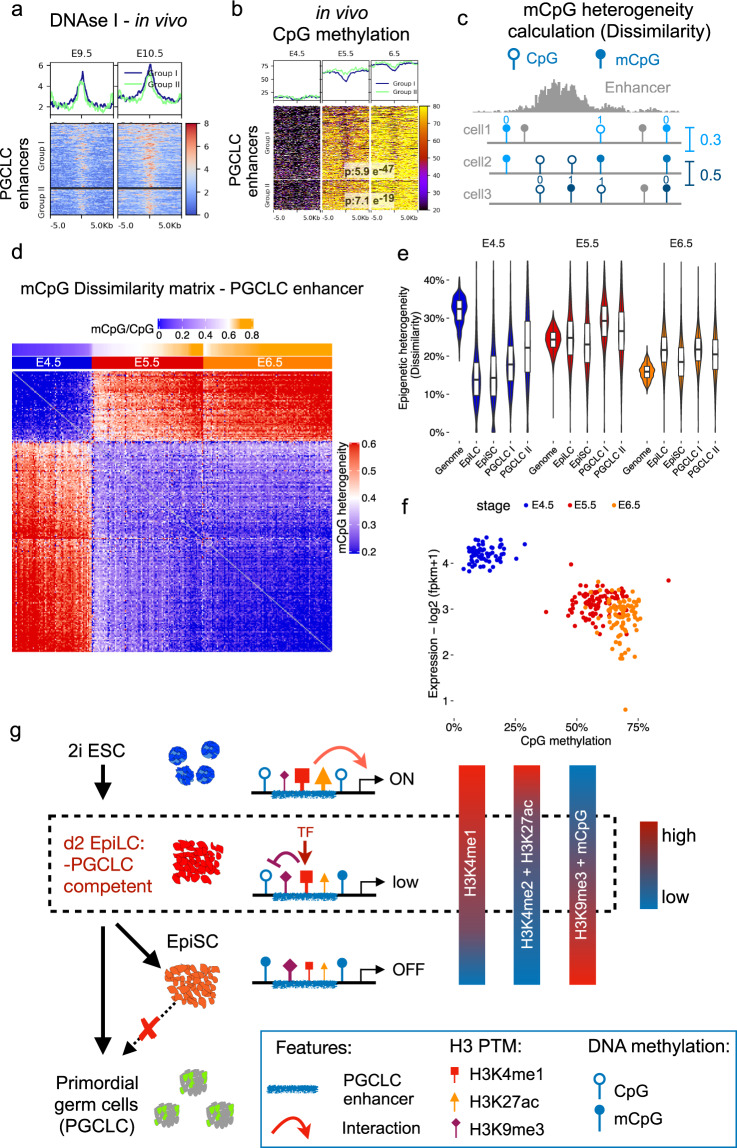
Fig. 7. Chromatin features of PGCLC enhancers in vivo.
a DNAse-seq levels for Group I and II PGCLC enhancers in PGCs from E9.5 and E10.5 mouse embryos67. Scales are in RPGC. b CpG methylation levels for Group I and II PGCLC enhancers in E4.5, E5.5, and E6.5 mouse epiblasts68. P-values were calculated using paired two-sided Wilcoxon tests. Scales: percentage of methylated CpGs. c mCpG heterogeneity was estimated based on the mCpG dissimilarity concept. For each pairwise comparison, the methylation status of CpGs covered in the two cells being compared is considered (blue lollipops). If two cells show the same or different methylation status they receive a value of 0 (similarity) or 1 (dissimilarity), respectively. The mean of all pairwise comparisons reflects CpG methylation heterogeneity. d Heatmap showing mCpG heterogeneity within PGCLC enhancers between pairs of individual cells. mCpG heterogeneity values are presented with a blue-red scale (blue: similar; red: dissimilar). The developmental stages of the investigated cells (E4.5, E5.5, or E6.5) and the average CpG methylation (blue-orange scale) measured for all PGCLC enhancers within each cell (n = 261 cells) are shown above the heatmap. e CpG methylation heterogeneity was measured in E4.5, E5.5, and E6.5 epiblast cells for all covered CpGs in the mouse genome as well as within EpiLC, EpiSC, and PGCLC enhancers. The horizontal lines in the boxplots indicate the median, the boxes the first and third quartiles, and the whiskers the ± 1.5 × interquartile range. f Comparison of the single-cell CpG methylation of Group I PGCLC enhancers and the single-cell RNA expression of genes linked to them (n = 256) in E4.5, E5.5, and E6.5 epiblasts. fpkm: Fragments per kilobase per million mapped reads. g Model illustrating the potential relevance of the partial decommissioning of Group I PGCLC enhancers for in vitro germline competence. The persistence of H3K4me1 within PGCLC enhancers in EpiLC might protect them from heterochromatinization (H3K9me3 and CpG methylation) and render them accessible and responsive to transcriptional activators. In turn, this can confer germline competence by facilitating PGCLC enhancer (re)activation during PGCLC differentiation.