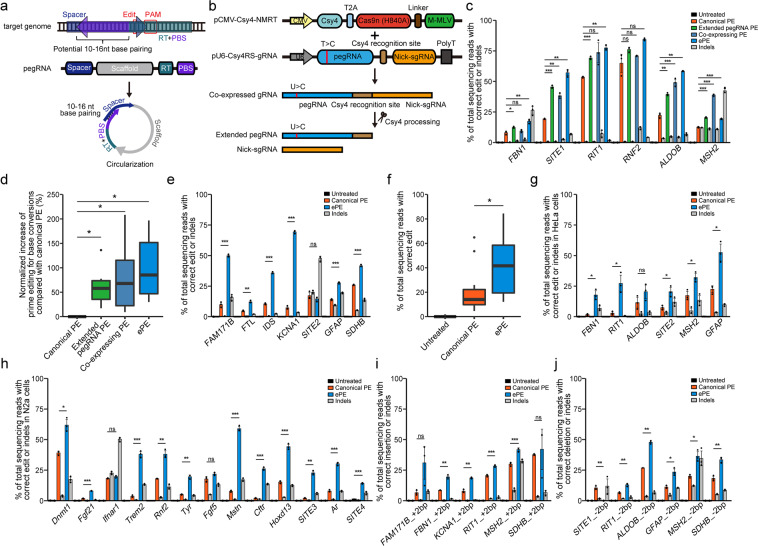
Fig. 1. Enhanced prime editing system using Csy4-processed pegRNA.
a A schematic representation of the circularization formed by the PBS and spacer. A canonical pegRNA consists of spacer, scaffold, RT, and PBS. pegRNA spacer is highlighted in dark blue, scaffold in gray, RT in cyan, PBS in purple. The spacer and the PBS share a complementary sequence, and their annealing is expected to cause pegRNA circularization. b ePE system. Csy4 protein is fused to and co-expressed with the prime editor NMRT. pegRNA and nick-sgRNA are fused and co-expressed in a single transcript from a U6 promoter with pegRNA flanked by Csy4 recognition site (Csy4RS). Csy4 nuclease cleaves and releases pegRNA and nick-sgRNA from the transcript. With Csy4 processing, the hairpin Csy4 recognition site remains at the 3′ end of the pegRNA to become extended pegRNA. Mutation of the fourth uracil of consecutive uracils (marked by red line) was introduced to the scaffold of pegRNA. c Increasing targeted efficiency of base transition and transversion by extended pegRNA, co-expressing extended pegRNA and nick-sgRNA, and ePE system in HEK293T cells. PCR amplicons from the target regions were analyzed by targeted deep sequencing. The reads only harboring correct edit were counted to evaluate the editing efficiency, and the reads harboring any undesired insertion or deletion were counted to evaluate the indel frequency. Gray bar indicates the indel frequency coupled with the editing efficiency indicated by the left closest bar. d Statistical analysis of normalized increase of targeted base transition and transversion editing efficiencies in c. e ePE system increases targeted efficiency of base transition and transversion at more sites in HEK293T cells. Gray bar indicates the indel frequency coupled with the editing efficiency indicated by the left closest bar. f Statistical analysis of prime editing point mutation efficiency by canonical PE and ePE system at all human sites used in c and e. g ePE system increases targeted efficiency of base transition and transversion in HeLa cells. Gray bar indicates the indel frequency coupled with the editing efficiency indicated by the left closest bar. h ePE system increases targeted efficiency of base transition and transversion in murine N2a cells. Gray bar indicates the indel frequency coupled with the editing efficiency indicated by the left closest bar. i ePE system increases the efficiency of targeted precise sequence insertion in HEK293T cells. Gray bar indicates the indel frequency coupled with the editing efficiency indicated by the left closest bar. j ePE system increases the efficiency of targeted precise sequence deletion in HEK293T cells. Gray bar indicates the indel frequency coupled with the editing efficiency indicated by the left closest bar. For c–j, data are presented as mean values ± SD, n = 3 independent experiments, two-tailed Student’s t-test (*P < 0.05, **P < 0.005, ***P < 0.0005).