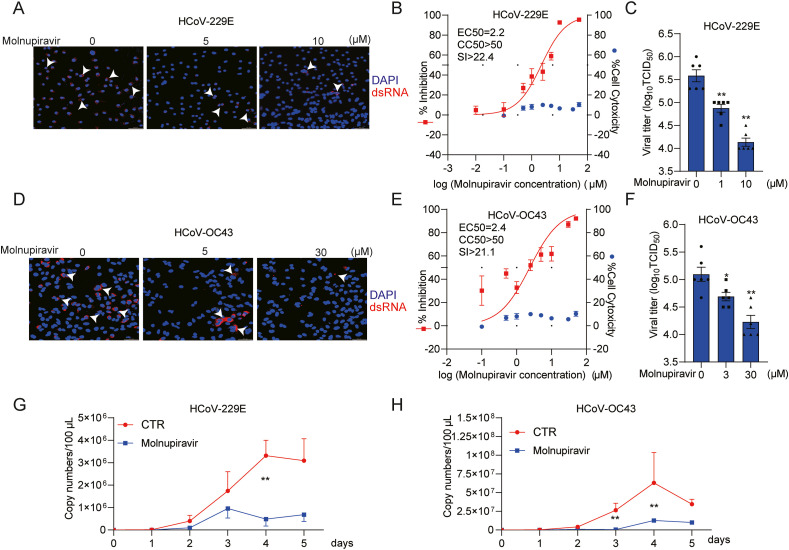
Fig. 3.
Antiviral effects of molnupiravir against HCoV-229E and HCoV-OC43 infection. (A) and (D) Immunofluorescence microscope analysis of dsRNA (red) upon treatment with different concentrations of molnupiravir in A549 cell line. Nuclei were visualized by DAPI (blue). (B) and (E) A549 cells were infected HCoV-229E or HCoV-OC43 at an MOI of 0.1 in the treatment of different concentrations of molnupiravir for 48 h. The viral yield in the cell supernatant was then quantified by qRT-PCR. Cytotoxicity was determined by MTT assay. The half maximum effective concentration (EC50) and the half maximum cytotoxic concentration (CC50) were calculated based on the model Y = 100/(1 + 10^(LogEC50-X)) using GraphPad Prism 8.0.2 software. The left and right Y-axis of the graphs represent mean % inhibition of virus yield and cytotoxicity of the drugs, respectively. (n = 6–16). (C) and (F) The supernatant and cells of each well under molnupiravir treatment was harvested after freezing and thawing for three times. Virus titers from different groups were determined by TCID50 assay (n = 6). (G) and (H) A549 cells were infected with HCoV-229E or HCoV-OC43 at the MOI of 0.1, and then untreated or treated with 5 μM molnupiravir for 5 days. Supernatant was collected every day to quantify secreted viruses by qRT-PCR, calculated as genomic copy numbers (n = 6). Standard curve for calculation of genomic copy numbers is included in Supplementary Figs. S3B and S3C. Data represent as mean ± SEM. *P < 0.05; **P < 0.01; ***P < 0.001. HCoV: human coronavirus. (For interpretation of the references to color in this figure legend, the reader is referred to the Web version of this article.)