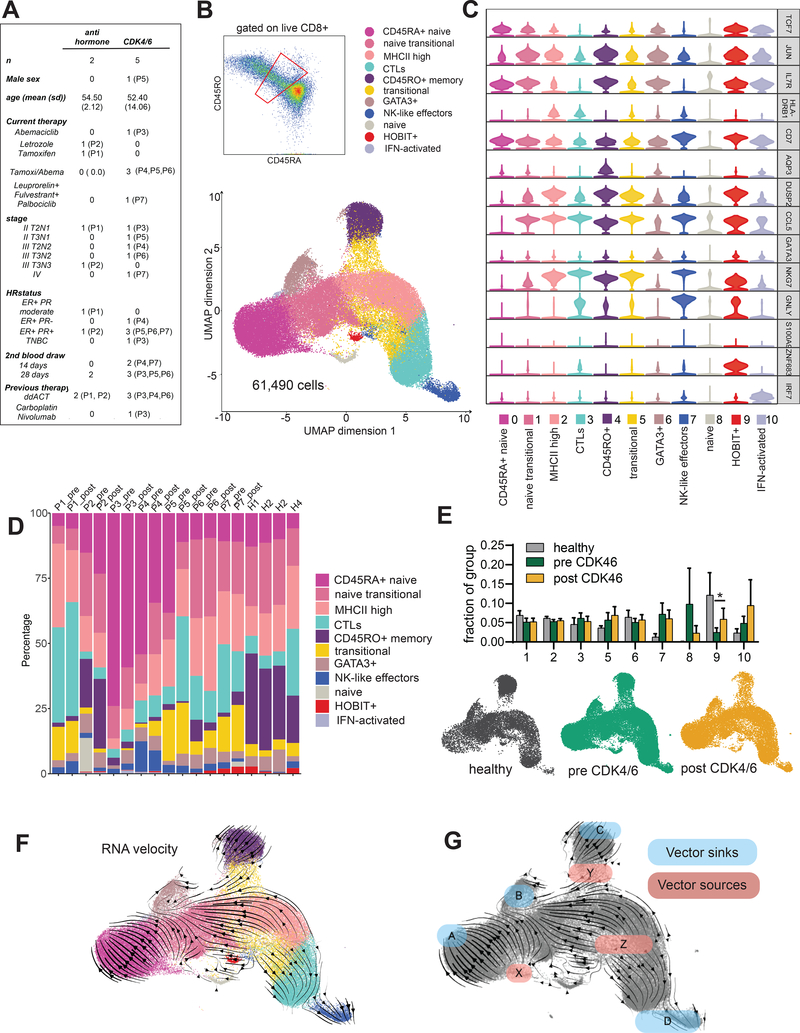
Figure 5: Memory cell precursors can be found by single cell transcriptional analysis of recently activated CD8 T cells from human peripheral blood.
A) Clinical characteristics of the patient cohort. B) Peripheral blood mononuclear cells were enriched for CD8 T cells using magnetic beads and stained with antibodies to CD8, CD45RO, and CD45RA. Cells were gated on live CD8+ prior to sorting the population gated in red. Single cell transcriptional profiling was performed using the 10X Genomics platform. 61,889 cells were analyzed from a combined 4 healthy donors and paired pre- and on-treatment samples from the 7 breast cancer patients shown in A. Cells were projected onto 2 dimensions by UMAP and grouped into 11 clusters. C) Violin plots of select cluster defining genes. D) Cluster representation per sample. E) Fractional representation of healthy, pre CDK4/6 and post CDK4/6 cells represented in each cluster. CD45RA+ naïve cells (cluster 0) and CD45RO+ memory cells (cluster 4) were excluded from the analysis. Error bars are SEM. Significance was evaluated using paired analysis of pre-CDK4/6 and post-CDK4/6 values for each patient; only cluster 9 was significant, *p=0.0403. UMAP projections of cells from healthy, pre and post CDK4/6 samples are shown. P1 and P2 post samples are included in the pre-CDK4/6 plot. F) RNA velocity analysis. G) Diagram of sinks and sources based on RNA velocity.